Lumpy Skin Disease Virus: Molecular Detection and Outbreak Investigation among Cattle in Gursum District of Somali Regional State, Ethiopia
| Received 13 Aug, 2024 |
Accepted 18 Dec, 2024 |
Published 30 Jun, 2025 |
Background and Objective: In August 2020, a lumpy skin disease outbreak affected cattle in the Gursum District of the Ethiopian Somali Regional State. According to the report from districts’ animal health professionals and officers, the outbreak killed several cattle and endangered many more. Thus, this study was designed to conduct both an epidemiological assessment and molecular diagnosis of the suspected LSD outbreak in the Gursum District. Materials and Methods: A cross-sectional study was conducted at Gursum District from September, 2020 to February, 2021, for epidemiological assessment and molecular diagnosis of the suspected LSD outbreak. Twelve skin-scraped nodule samples were purposively collected from clinically diseased cattle and sixty animal owners were interviewed in six kebeles. Virus isolation was performed on Vero cell lines and molecular detection was performed on classical Polymerase Chain Reaction (PCR) to confirm the LSD virus. In addition, the Chi-square (χ2) test and frequency measurements were used for analyzing and presenting the epidemiological data interviewed from animal owners. Results: All samples showed the LSD virus’s cytopathic effects. The PCR detected the LSD viral DNA and confirmed the cause of the suspected LSD outbreak. According to the epidemiological investigation and questionnaire survey from respondents, the disease had an overall mortality and morbidity rate of 12.52% (74/591) and 33.67% (199/591), respectively. Furthermore, on Chi-square analysis; the age of cattle, season and contact at the communal point were statistically significant (p<0.05) indicating the major factors for the outbreak occurrence of the disease. The findings of molecular analysis confirmed that the outbreak was caused by LSD virus and epidemiological factors worsened the disease. Conclusion: Thus, regular vaccination of cattle at trafficking areas, communal points and watering areas should be carried out to reduce the disease.
| Copyright © 2025 Bitew et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Lumpy skin disease (LSD) is a contagious re-emerging transboundary and notifiable viral disease of cattle and buffaloes causing significant economic losses in livestock production1. Originally only affecting Africa, the disease spread from the Middle East to South East Europe in 2012, affecting EU Member States (Greece and Bulgaria) and numerous other Balkan nations2. Despite the typically low death rate, there are considerable financial losses as a result of condition losses, decreased milk supply, abortions, infertility and damaged hides3. Sporadic or epizootic occurrences of lumpy skin disease are possible and they are more prevalent in warm and humid climates. In endemic regions, incidences usually increase in the warmer, wet months when there are more insect vectors and decrease in the dry months. When incursion takes place in European cattle breeds, mortality and morbidity can both be significantly higher (morbidity occasionally reaching 100%)4,5.
The disease is caused by the lumpy skin disease virus which belongs to the genus Capripoxvirus6. Conically inverted necrotized skin nodules together with swollen lymph nodes, a persistent fever, lacrimation, nasal discharge, pneumonia and occasionally mortality, are the hallmarks of lumpy skin disease7. Although clinical cases in Asian water buffalo (Bubalus bubalis) have also been reported, the disease primarily affects cattle and sheep and goats appear unaffected even when they are close to cattle during outbreaks8.
Lumpy Skin Disease Virus (LSDV) is thought to be transmitted primarily by arthropod vectors. Mosquitoes, biting flies (e.g., Stomoxys calcitrans and Biomyia fasciata), culicoides midges and hard ticks (e.g., Amblyomma hebraeum and Rhipicephalus spp.) are currently thought to be mechanical vectors. Ticks probably play little or no role when the LSD virus is spreading rapidly during epizootics; however, they might be involved in transmission and maintenance in endemic regions. Prolonged virus survival has been reported in some vectors. The most important vector is likely to vary between affected regions, depending on the climate, season, environmental temperature, humidity and vegetation, favoring different insect and tick species9,10.
Lumpy skin disease viruses can be detected via virus isolation, electron microscopy, serological and molecular techniques9. Furthermore, LSD virus antigens have been found using Restriction Fragment Length Polymorphism (RFLP)11, which has also been used to distinguish virulent LSD viruses from vaccination strains12.
Lumpy skin disease is currently endemic in most of Africa, parts of the Middle East and Turkey. In Ethiopia, LSD was first observed in 1983 in the western and north-western parts of the country13. The disease has now spread to almost all regions and agroecological zones of the country14. According to the report from Ayelet et al.14, several LSD outbreaks were reported in Ethiopia from 2007-2011. The five-year retrospective analysis of the study showed that a total of 1,675 outbreaks with 62,176 cases and 4,372 deaths of cattle were reported. The highest number of outbreaks were reported in Oromia (1,066), followed by Amhara (365) and the Southern Nations, Nationalities and People’s Region (123) causing huge cattle losses.
The main livestock production systems of the Somali Regional States (SRS) are pastoral and agro-pastoral and the society’s sole source of income is mainly cattle products15. The occurrence of an outbreak of notifiable diseases such as LSD could have a great economic impact on society. In late August, 2020, there was a report of a suspected LSD outbreak in the Gursum District of the Ethiopian Somali Regional State. According to the report from district animal health professionals and officers, the outbreak affected and killed several cattle and also put thousands at risk. Thus, prompt a molecular diagnosis of the suspected disease that necessary to identify the etiological agent and questionnaires to assess the intensity and associated factors of the disease for effective and timely intervention. Hence, this study attempted to undergo molecular diagnosis and epidemiological investigation of suspected LSD outbreaks in the Gursum District.
MATERIALS AND METHODS
Study area: The study was carried out in the Gursum District, which is situated 600 km East of Addis Ababa, Ethiopia, in the Fafan/Jigjiga/Administrative Zone of the Somali Regional State. The zone is located between roughly 9020' North East and 45,056' East, with an elevation difference of 500-1650 m above sea level. The high temperatures are indicative of a semi-arid climate. In the area, the average yearly rainfall is between 600 and 700 mm. It comprises eleven districts and is best exercised with agro-pastoralism 56.8%, followed by pastoralism and sedentary production systems comprising 34.1 and 9.1%, respectively. It has a total of 5.58 million cattle, 0.8 million sheep, 0.7 million goats and 0.13 million camel population16.
Study design: A cross-sectional study was conducted from September, 2020 to February, 2021 for epidemiological assessment and molecular diagnosis of the suspected LSD outbreak in the Gursum District on cattle manifesting typical clinical signs of the disease.
Sampling methods and sample collection: In August 2020, the College of Veterinary Medicine of Jigjiga University received a notification regarding a suspected outbreak of LSD in the Gursum District of Fafan Zone, Somali Regional State. The outbreak happened in six Kebeles of the Gursum District namely in Tikdem, El-halrad, Fafem, Halago, Kuda and Adada. For the questionnaire survey, sixty cattle owners from whom their cattle were affected by the LSD were selected from the respective kebeles purposively.
Twelve representative clinical samples (skin nodules) were collected from animals manifesting typical clinical signs of the disease. The samples were collected aseptically by scraping from characteristic skin nodules and placed in sterile universal bottles with a transport medium (phosphate-buffered solution) containing an antibiotic-penicillin. All samples were securely capped, labeled with permanent markers and kept in a cool box that had cooling elements. The samples were immediately transported to the National Veterinary Institute (NVI) for processing and molecular analysis.
Laboratory analysis
Propagation of cell line: The Vero cell line monolayer was maintained in 25 cm3 disposable tissue culture flasks. All the operations of harvesting, seeding and infection of the cells were performed in laminar airflow (Scitek Global, Houston, United States), in a sterile environment. Cells were diluted in a growth medium to obtain approximately 1×105/mL and distributed in 25 cm3 disposable tissue culture flasks. As 5 to 7 mL of growth medium was added per tissue culture flask. The seeded flask was incubated at 37°C in a 5% CO2 incubator (Scitek Global, Houston, United States) till the monolayer was complete. After 48 hrs of incubation, the growth medium was discarded and 5 to 7 mL of maintenance medium was added to the flask without disturbing the monolayer as described by Freshney17.
Virus isolation: The nodule scrapping samples were thawed at room temperature and washed three times using a sterile phosphate-buffered solution that contains an antibiotic under laminar airflow (Scitek Global, Houston, United States) hood class II. About 1 g of each sample was grounded using a sterile mortar and pestle by adding 9 mL of sterile phosphate buffered solution (pH, 7.2) containing antibiotic. The tissue suspension was centrifuged at 3500 rpm for 10 min. The supernatant was collected and about 1 mL of tissue suspension was inoculated on a fully grown monolayer Vero cell line grown on 25 cm3 tissue culture flasks and incubated for 1 hr at 37°C then, the flask was flashed with 10 mL of the suitable medium of Glasgow Eagle Minimum Essential Medium (GMEM), containing antibiotic and 2% calf serum and incubated at 37°C. Cells were monitored daily for 14 days for the occurrence of virus-induced cytopathic effects (CPE) under inverted microscope (CHD Medical, Chicago, USA), with changing the medium as needed18.
Viral DNA extraction: The viral DNA was extracted from tissue culture supernatant using Dneasy® blood and tissue DNA extraction kit (QIAGEN, USA) following manufacturers’ instructions. In brief, 200 μL tissue suspension supernatant was mixed with 20 μL proteinase K enzyme and 200 μL lysis buffer and incubated at 56°C in a water bath for 30 min. As 200 μL pure ethanol alcohol (98%) was added to the mixture and transferred to the DNeasy minispin column. The DNeasy minispin column was placed in to collection tube and centrifuged at 125 rpm for 90 sec. As 500 μL washing buffer was added and centrifuged at 125 rpm for 90 sec the fluid through in the collection tube was discarded and the minispin column was transferred to another collection tube. As 500 μL concentrated washing buffer was added and centrifuged at 134 rpm for 3 min at which, the filtered fluid was discarded and the DNeasy minispin column was centrifuged at 134 rpm for 2 min leaving the centrifuge open. Finally, 50 μL elution buffer was added and centrifuged at 134 rpm for 3 min by placing the filtrated minispin column into a 1.5 mL Eppendorf tube. The extracted DNA was transported to the DNA amplification room after discarding the minispin column.
Polymerase chain reaction-classical PCR: Classical PCR amplification and detection of the extracted DNA samples were performed in 2720 thermal cycler (Applied Biosystems) using primers of SPGPRNApol-Fow-5 pm/μL-5'-TCTATGTTCTTGATATGTGGTGGTAG-3' and SPGPRNApol-REV-5 pm/μL-5'-AGTGATTAGGTGGTGTATTATTTTCC-3' described by NVI (NVI, Unpublished observations). The PCR reaction was carried out in a total volume of 20 μL containing, 2 μL forward primer, 2 μL reverse primer, 10 μL IQ super mix, 3 μL template DNA and 3 μL of RNase-free water. The thermal cycling conditions included an initial step at 95°C for 5 min, which was followed by 40 cycles of denaturation at 95°C for 30 sec, annealing at 50°C for 30 sec and elongation at 72°C for 30 sec as well as a final extension step at 72°C for 7 min.
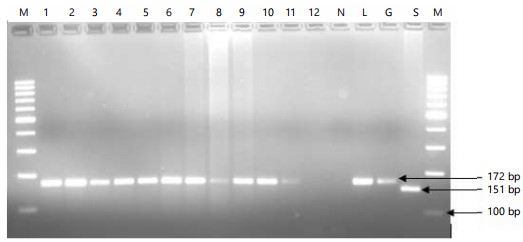
Amplified results were examined using agarose gel electrophoresis, employing a Gen Ruler TM 100 bp Plus DNA ladder as a molecular marker on a 3% agarose gel prepared in Tris/Acetate/EDTA (TAE) buffer. A mixture of 10 μL PCR product and 4 μL loading buffer containing loading dye was added to the wells of a gel that had already been prepared. The gel was then run at 120 volts for approximately 1.2 hrs in parallel with the DNA molecular marker (ladder) in the electrophoresis apparatus until the DNA samples had sufficiently migrated through the gel. The size of the bands that developed on an agarose gel was used to validate positive results after the DNA bands were observed using a UV transilluminator (BioDoc-ItTM, California, USA) at a wavelength of 590 nm. The 172 bp product of PCR was found to contain LSD virus DNA, which was determined to be a positive result.
Questionnaire survey: A team of experts was dispatched to the area and thoroughly observed cases on the ground, spoke to members of the communities and gathered the necessary information to make a rapid appraisal and ground-level situation analysis of the case. A semi-structured questionnaire was prepared and cattle owners were interviewed. A total of 60 owners were involved from 6 kebeles; 10 respondents in each kebele and interviewed about their herd dynamics, the numbers, age and sex of cattle affected, vaccination status, the number of dead cattle and seasonal aspects of the diseases.
Data management and analysis: Collected data were encoded and analyzed using Statistical Package for Social Sciences (SPSS) software version 20. The Chi-square (χ2) was held to test the presence of association among different categorized variables. In all the analysis, confidence levels at 95% were calculated and p<0.05 was used as a statistically significant19. In addition, descriptive statistics were used to quantify the results on theawareness of the respondents about the disease, age and sex of the affected and dead cattle during the outbreak.
Ethical approval
Ethics approval and consent to participate: All the study activities in this work were conducted based on ethical standards after ethical approval from the Jigjiga University Research Ethics Review Committee Reg. No. (JJU-RERC 050/2020). Investigators treated animals with kindness and took proper care by minimizing discomfort, distress or pain. They assumed that all procedures that would cause pain in human beings may cause pain in study animals. In addition, Owners’ consent of their interest was taken verbally for sampling and data collection.
RESULTS
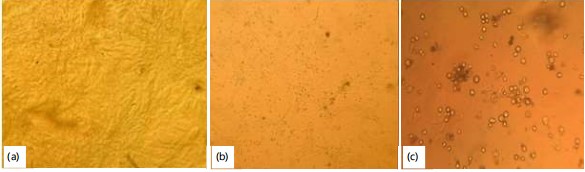
The infected animals showed typical LSD clinical signs; intradermal nodules on the skin, fever, depression, lacrimation, ulcerative skin lesions and recumbency (Fig. 1).
Virus isolation: The inoculated suspension samples were observed for the formation of cytopathic effects (CPE) by comparing parallel to the control Vero cell line under an inverted microscope. The cytopathic effects were observed starting on the 5th day post-inoculation in the first passage. Accordingly, viral isolation from collected samples on Vero cells revealed that all the samples (n = 12,100%) showed CPE. The CPE was characterized by monolayer destruction, rounding of infected cells which was formed singly and aggregation of dead cells. None of the negative controls produced any CPE (Fig. 2).
Classical PCR: From a total of 12 samples processed, 11 samples resulted in an amplification of the expected size of the amplicon (172 bp) indicating the presence of the LSD virus genome. However, 1 sample produced a faded band at the site of the expected amplicon size (Fig. 3).

|
|
|
| Table 1: | Mortality, morbidity and case fatality rates of lumpy skin disease in cattle from kebeles during the outbreak | |||
| Kebeles | Number of susceptible cattle |
Number of affected |
Number of death |
Morbidity rate (%) |
Mortality rate (%) |
Case fatality rate (%) |
| Tikdem | 192 | 66 | 21 | 34.37 | 10.93 | 31.81 |
| El-halrad | 62 | 19 | 4 | 30.64 | 6.45 | 21.05 |
| Fafen | 189 | 51 | 23 | 26.98 | 12.17 | 54.09 |
| Halago | 53 | 12 | 5 | 22.64 | 9.43 | 41.67 |
| Kuda | 48 | 21 | 12 | 43.75 | 25 | 57.14 |
| Adad | 47 | 30 | 9 | 63.82 | 19.14 | 30.00 |
| Total | 591 | 199 | 74 | 33.67 | 12.52 | 37.18 |
| Table 2: | Phenomenon of LSD outbreak in relation to factors; age and sex of cattle | |||
| Factor | Number of susceptible |
Number of affected |
Number of died |
Morbidity rate (%) |
Mortality rate (%) |
Chi-square | p-value | Case fatality rate (%) |
| Age | ||||||||
| Young <2 years | 276 | 116 | 48 | 42.02 | 17.39 | 16.194 | 0.001 | 41.38 |
| Adult >2 years | 315 | 83 | 26 | 26.34 | 8.25 | - | - | 31.32 |
| Sex | ||||||||
| Male | 240 | 80 | 25 | 33.33 | 10.41 | 0.021 | 0.886 | 31.25 |
| Female | 351 | 119 | 49 | 33.9 | 13.96 | - | - | 41.17 |
| Total | 591 | 199 | 74 | 33.67 | 12.52 | - | - | 37.18 |
Questioner survey: According to the questionnaire, the pastoralists named the LSD after a local name “kuskus” and showed the significance of the diseases with associated risk factors for the outbreak. About 53 of the 60 respondents were aware of LSD from previous outbreaks that last occurred in two years, while 7 respondents developed awareness from observing their diseased cattle during the outbreak. During the investigation, LSD recorded overall mortality (12.52%), morbidity (33.67%) and case fatality (37.18 %) rates and the highest mortality rates occurred in Kuda 25%, followed by Adad 19.14% and Fafan 12.17% (Table 1).
The effect of the diseases on animal sex showed that higher mortality (13.96%) and case fatality (41.15%) rates were recorded in female than male cattle 10.41 and 31.25%, respectively. A higher mortality (17.39%) was observed in young than adult cattle (8.25%) (Table 2). The clinical presentation of LSD was significantly higher (p<0.05) in young cattle (p = 0.001), cattle contact at communal points; watery and grazing areas (p = 0.035) and in the rainy season (p = 0.001) (Table 3).
| Table 3: | Summary of LDS outbreak associated risk factors acquired from questionnaire survey participants | |||
| Respondents | ||||||
| Factor | Number | Percentage | Number of susceptible cattle |
Number of affected cattle | Chi-square | p-value |
| Vaccination | ||||||
| No | 45 | 75 | 460 | 163 | 2.888 | 0.089 |
| Yes | 15 | 25 | 131 | 36 | ||
| Likely source of outbreak | ||||||
| Communal points | 35 | 58.3 | 353 | 132 | 6.737 | 0.035 |
| Cattle movement | 20 | 33.3 | 164 | 50 | ||
| Introduction of new animals | 5 | 8.3 | 74 | 17 | ||
| Season | ||||||
| Rainy | 55 | 91.6 | 483 | 178 | 11.977 | 0.001 |
| Dry | 5 | 8.3 | 108 | 21 | ||
| Total | 60 | 591 | 199 | |||
DISCUSSION
In the current study, the lumpy skin disease virus was identified as the cause for cattle exhibiting clinical signs of; intradermal nodules on the skin, fever, depression, lacrimation and ulcerative skin lesions. This finding agreed with Alemayehu et al.20, who reported on different parts of the country as cattle affected with a lumpy skin disease were manifesting such clinical signs. On cell culture, all the samples (n = 12) were positive for virus isolation, which is indicated by a CPE of rounding of single cells, aggregation of dead cells and destruction of the monolayer. This result was in agreement with Tassew et al.10 and Ayelet et al.14, who performed virus isolation on Vero cells from tissue samples of LSD in Ethiopia. In this study, the results of 11 skin-scraped samples were indicative of the LSD virus having an amplicon size of l72 bp. Compatible findings were reported by Tassew et al.10, Gebeyehu et al.21 and Gari et al.22, that LSDV was conformed from tissue samples using conventional PCR in different parts of Ethiopia.
The current outbreak of LSD occurred from July to October. These months were characterized by moderate to heavy rain in the area that encouraged the growth and distribution of insects and was possibly linked to an increase in insect vector occurrence. This could probably exacerbate LSD virus transmission by infected arthropods from infected to naïve cattle. This finding was in agreement with the reports of Grassly and Fraser23 and Molla et al.24. The other important issue in deciding the occurrence and distribution of the disease were interviewing cattle owners about what would be the likely source of an outbreak of the disease in the study kebeles, as owners (n = 35, 58.3%) responded that, contact at communal points like watering and grazing pastures would be considered as the major factor for the dissemination of the disease to the neighboring kebeles, this agreed with the study of Selim et al.25, Hailu et al.26 and Tuppurainen et al.27.
In the current study, a higher morbidity rate (33.67%) was recorded than that reported by Alemayehu et al.20 (13.61%) and Leliso et al.28 (18%). This high frequency of the disease’s dissemination may probably be due to cattle in pastoral areas being spent traveling long distances searching for feed and water, in addition to sharing common grazing pasture and watering points. Similarly, the current mortality rate of the disease (12.52%) is relatively higher than studies reporting a 10% mortality rate7, 4.97%14, 0.99%29 and 0.50%30. This might be due to variations in the immune status of the animals, the pathogenic nature of circulating virus strains and breeds of cattle. Concerning ages, young cattle had a higher morbidity rate (42.02%) than adult cattle (26.34%), it is agreed with Ayelet et al.14; 18 and 11.9%. This may probably be due to the weak immunity status of young cattle with low or no exposure to the LSD virus from previous infections.
CONCLUSION
The present study confirmed that the suspected outbreak in the Gursum District was caused by the LSD virus through virus isolation and classical PCR assay. The semi-aired nature of pastoral areas; watering pools and scarce grazing lands, are making this disease more devastating and resulting in high cattle losses that put in danger for the livelihood of pastoralists and agro-pastoralists. Thus, prompt emergency immunization of cattle and increasing cattle watering pools with restriction of cattle movement for long distances should be implemented in the area. In addition, further characterization of the detected Lumpy skin disease virus and diagnosis of LSD in other parts of Ethiopia is required.
SIGNIFICANCE STATEMENT
This study investigates a lumpy skin disease (LSD) outbreak in the Gursum District of Ethiopia, in August 2020, which significantly impacted the cattle population in the area. Through a cross-sectional design, twelve skin-scraped samples were analyzed and sixty animal owners were interviewed to assess the outbreak’s epidemiology. Results indicated a mortality and morbidity rate of 12.52 and 33.67%, respectively. Molecular diagnosis confirmed the presence of the LSD virus and factors such as cattle age, seasonal variations and communal contact points were significantly influencing disease prevalence. The findings underscore the urgent need for any means of reducing animal contact and conducting regular vaccinations in high-risk areas to mitigate future outbreaks and protect livestock health.
ACKNOWLEDGMENT
The authors would like to thank the Ethiopian National Veterinary Institute and animal owners who rendered this work during the study period.
REFERENCES
- Sudhakar, S.B., N. Mishra, S. Kalaiyarasu, S.K. Jhade and D. Hemadri et al., 2020. Lumpy skin disease (LSD) outbreaks in cattle in Odisha State, India in August 2019: Epidemiological features and molecular studies. Transboundary Emerging Dis., 67: 2408-2422.
- Kiplagat, S.K., P.M. Kitala, J.O. Onono, P.M. Beard and N.A. Lyons, 2020. Risk factors for outbreaks of lumpy skin disease and the economic impact in cattle farms of Nakuru County, Kenya. Front. Vet. Sci., 7.
- Roche, X., A. Rozstalnyy, D. TagoPacheco, C. Pittiglio and A. Kamata et al., 2020. Introduction and Spread of Lumpy Skin Disease in South, East and Southeast Asia: Qualitative Risk Assessment and Management. 1st Edn., Food and Agriculture Organization of the United Nations, Rome, Italy, ISBN: 978-92-5-133563-5, Pages: 62.
- Tuppurainen, E.S.M. and C.A.L. Oura, 2012. Review: Lumpy skin disease: An emerging threat to Europe, the Middle East and Asia. Transboundary Emerging Dis., 59: 40-48.
- Pestova, Y.E., E.E. Artyukhova, E.E. Kostrova, I.N. Shumoliva, A.V. Kononov and A.V. Sprygin, 2018. Real time PCR for the detection of field isolates of lumpy skin disease virus in clinical samples from cat-tle. Agric. Biol., 53: 422-429.
- Tulman, E.R., C.L. Afonso, Z. Lu, L. Zsak, G.F. Kutish and D.L. Rock, 2001. Genome of lumpy skin disease virus. J. Virol., 75: 7122-7130.
- Salib, F.A. and A.H. Osman, 2011. Incidence of lumpy skin disease among Egyptian cattle in Giza Governorate, Egypt. Vet. World, 4: 162-167.
- Mulatu, E. and A. Feyisa, 2018. Review: Lumpy skin disease. J. Vet. Sci. Technol., 9.
- Tuppurainen, E.S.M., E.H. Venter, J.L. Shisler, G. Gari and G.A. Mekonnen et al., 2017. Review: Capripoxvirus diseases: Current status and opportunities for control. Transboundary Emerging Dis., 64: 729-745.
- Tassew, A., A. Assefa, E. Gelaye, B. Bayisa and M. Ftiwi, 2018. Identification and molecular characterization of lumpy skin disease virus in East Hararghe and East Shoa Zone, Oromia Regional State. ARC J. Anim. Vet. Sci., 4: 1-16.
- Yousefi, P.S., K. Mardani, B. Dalir-Naghadeh and G. Jalilzadeh-Amin, 2017. Epidemiological study of lumpy skin disease outbreaks in North-Western Iran. Transboundary Emerging Dis., 64: 1782-1789.
- Menasherow, S., M. Rubinstein-Giuni, A. Kovtunenko, Y. Eyngor and O. Fridgut et al., 2014. Development of an assay to differentiate between virulent and vaccine strains of Lumpy Skin Disease Virus (LSDV). J. Virol. Methods, 199: 95-101.
- Mebratu, G.Y., B. Kassa, Y. Fikre and B. Berhanu, 1984. Observation on the outbreak of lumpy skin disease in Ethiopia. J. Livest. Vet. Med. Trop. Countries, 37: 395-399.
- Ayelet, G., R. Haftu, S. Jemberie, A. Belay and E. Gelaye et al., 2014. Lumpy skin disease in cattle in Central Ethiopia: Outbreak investigation and isolation and molecular detection of the virus. Sci. Tech. Rev., 33: 877-887.
- Birhan, M., 2013. Livestock resource potential and constraints in Somali Regional State, Ethiopia. Global Vet., 10: 432-438.
- Zerihun, S., M. Mulugeta and M. Abi, 2024. Rural household resilience to food insecurity in Majang Zone, Southwestern Ethiopia. F1000Research, 13.
- Freshney, R.I., 2011. Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications. 6th Edn., John Wiley & Sons, Hoboken, New Jersey, ISBN: 9780470649350, Pages: 768.
- Mangana-Vougiouka, O., P. Markoulatos, G. Koptopoulos, K. Nomikou, N. Bakandritsos and P. Papadopoulos, 2000. Sheep poxvirus identification from clinical specimens by PCR, cell culture, immunofluorescence and agar gel immunoprecipitation assay. Mol. Cell. Probes, 14: 305-310.
- Moulton, L.H., M.C. Wolff, G. Brenneman and M. Santosham, 1995. Case-cohort analysis of case-coverage studies of vaccine effectiveness. Am. J. Epidemiol., 142: 1000-1006.
- Alemayehu, G., G. Zewde and B. Admassu, 2013. Risk assessments of lumpy skin diseases in Borena bull market chain and its implication for livelihoods and international trade. Trop. Anim. Health Prod., 45: 1153-1159.
- Gebeyehu, A., M. Taye and R. Abebe, 2022. Isolation, molecular detection and antimicrobial susceptibility profile of Salmonella from raw cow milk collected from dairy farms and households in Southern Ethiopia. BMC Microbiol., 22.
- Gari, G., A. Waret-Szkuta, V. Grosbois, P. Jacquiet and F. Roger, 2010. Risk factors associated with observed clinical lumpy skin disease in Ethiopia. Epidemiol. Infect., 138: 1657-1666.
- Grassly, N.C. and C. Fraser, 2006. Seasonal infectious disease epidemiology. Proc. R. Soc. B, 273: 2541-2550.
- Molla, W., M.C.M. de Jong and K. Frankena, 2017. Temporal and spatial distribution of lumpy skin disease outbreaks in Ethiopia in the period 2000 to 2015. BMC Vet. Res., 13.
- Selim, A., E. Manaa and H. Khater, 2021. Seroprevalence and risk factors for lumpy skin disease in cattle in Northern Egypt. Trop. Anim. Health Prod., 53.
- Hailu, B., T. Tolosa, G. Gari, T. Teklue and B. Beyene, 2014. Estimated prevalence and risk factors associated with clinical Lumpy skin disease in North-Eastern Ethiopia. Preventive Vet. Med., 115: 64-68.
- Tuppurainen, E.S.M., W.H. Stoltsz, M. Troskie, D.B. Wallace and C.A.L. Oura et al., 2011. A potential role for ixodid (hard) tick vectors in the transmission of lumpy skin disease virus in cattle. Transboundary Emerging Dis., 58: 93-104.
- Leliso, S.A., F.D. Bari and T.R. Chibssa, 2021. Molecular characterization of lumpy skin disease virus isolates from outbreak cases in cattle from Sawena District of Bale Zone, Oromia, Ethiopia. Vet. Med. Int., 2021. https://doi.org/10.1155/2021/8862180
- Kasem, S., M. Saleh, I. Qasim, O. Hashim and A. Alkarar et al., 2017. Outbreak investigation and molecular diagnosis of lumpy skin disease among livestock in Saudi Arabia 2016. Transboundary Emerging Dis., 65: e494-e500.
- Chala, W., K. Adamu, H. Mohammed, G. Deresse, S. Tesfaye and E. Gelaye, 2024. Outbreak investigation, isolation, and molecular characterization of lumpy skin disease virus in cattle from North West Oromia Region, Ethiopia. Vet. Med. Int., 2024.
How to Cite this paper?
APA-7 Style
Bitew,
Z., Abera,
T., Bizuayehu,
F., Mohammed,
F. (2025). Lumpy Skin Disease Virus: Molecular Detection and Outbreak Investigation among Cattle in Gursum District of Somali Regional State, Ethiopia. Asian Journal of Biological Sciences, 18(2), 254-262. https://doi.org/10.3923/ajbs.2025.254.262
ACS Style
Bitew,
Z.; Abera,
T.; Bizuayehu,
F.; Mohammed,
F. Lumpy Skin Disease Virus: Molecular Detection and Outbreak Investigation among Cattle in Gursum District of Somali Regional State, Ethiopia. Asian J. Biol. Sci 2025, 18, 254-262. https://doi.org/10.3923/ajbs.2025.254.262
AMA Style
Bitew
Z, Abera
T, Bizuayehu
F, Mohammed
F. Lumpy Skin Disease Virus: Molecular Detection and Outbreak Investigation among Cattle in Gursum District of Somali Regional State, Ethiopia. Asian Journal of Biological Sciences. 2025; 18(2): 254-262. https://doi.org/10.3923/ajbs.2025.254.262
Chicago/Turabian Style
Bitew, Zemenu, Tsegalem Abera, Fanuel Bizuayehu, and Fahmi Mohammed.
2025. "Lumpy Skin Disease Virus: Molecular Detection and Outbreak Investigation among Cattle in Gursum District of Somali Regional State, Ethiopia" Asian Journal of Biological Sciences 18, no. 2: 254-262. https://doi.org/10.3923/ajbs.2025.254.262

This work is licensed under a Creative Commons Attribution 4.0 International License.



