Constituents of Prokaryotic Organisms for Production of Extracellular Enzymes in the Development of Immunological Process
Background and Objective: The mutation rate with complete genome sequences had spectra of microorganisms and cell suspensions moved by the actin network beneath the membrane within eukaryotic cells. The research was to study exoenzyme an extracellular enzyme secretion by cells and functions outside of the organism. Materials and Methods: Sequence factors studied were sample populations of bacteria with the same variance for alteration of the conversion of the genes and traits. These are processed through the Golgi apparatus and released into the cells. This was studied using different variances for irregularities in the culture. Results: It was found cytosol a portion of the cytoplasm containment within membrane organelles. This made up 70% of cell volume and a mixture of hyphae dissolution of cells. Eukaryotic base numbers for the strands of cytoplasm. This suggested the cell surround was not permeable for diffusion process occurrences. Conclusion: The gene micro-arrays had interactions with oligo cells. It concluded phase extraction was needed in sets and preconcentration. The surrounds of glycan structures synthesis had the presence of glycan in biological samples of immunologically based measurements of cells.
| Copyright © 2023 Solomon Ifeanyichukwu Ubani. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
In the literature, the mutation rate with complete genome sequences was the result of spectra of microorganisms and cell suspensions moved by an actin network beneath the membrane within eukaryotic cells. An exoenzyme was responsible for the remarkable spatiotemporal organization inhabited by processes and absorption for the capability of bacteria and archaea diversity and important biological activities.
According to Aanen and Debets1 the mutation rate was an important feature of the evolution of organisms. But were mostly found in high-density regions of prokaryotic and eukaryotic cells and were difficult to acquire for biological responsiveness. In the literature, for the study of the mutation rate the gene sequences were found in almost all the prokaryotic and eukaryotic organisms for prediction of the products2. This was an important factor for studies of the mutation rate of specimens.
Cytoplasm was the content of all the cells. This included cytosol (a gel substance), organelles (cell structure) and cytoplasmic inclusions3. This was a kind of exoenzyme secreted by a cell and functions outside the cell. This was produced by both prokaryotic and eukaryotic processes.
These features of sequences were essential for the identification and classification of microorganisms and cells. The complexity was found in the composition and structures of the spectrum.
In this study, proteomics used the method that used initially for the development and application of methods for gene traits and functions with gene sequences of the organism in the culture4. The idea behind the work stated there were three components of the cytoplasm had on observation leading feature of the cell, adhesion and deadhesion of the cell for movement of membrane for identification and Quantitative spectroscopy analysis of prokaryotic cells: Vegetative cells and spores5. Phages were found to be the most abundant organism and a feature of mostly prokaryotic existence. A bacteriophage-affected bacterium.
Each of these caused a motion by segments, prokaryotes displayed a spatial-temporal organization of processes in the cell6. The study relied on the performance of microscopy images. Microbial inhabited all environments and contained a vast biological diversity7. This facilitated the phylogenetic class8.
Most times the exoenzymes disintegrated in macro substances. This was important for the allowance of the constituent’s passage through the cell membrane and into the cell.
The study was to examine the transfer of microorganism spectrums and cell suspensions through the actin network in eukaryotic cells, revealing mutation rates with entire genome sequences. Exoenzymes were the purpose for determining the secretion by cells. The research was to understand the Oligo cells interaction effect by the exoenzyme with gene micro-arrays, requiring preconcentration and set phase extraction. The synthesis of glycan structures was to determine the effect of glycosylation in biological samples based on immunology.
MATERIALS AND METHODS
Study area: The study was carried out in Microbiology Department, Quality Control Lab, Egypt from January, 2018 to March, 2019 and in Association for Molecular Pathology in May, 2022.
Cellular participants for decomposition in the combination of functions of a gland. These were an acinar cell and duct cell for enzyme and secretion. The cytoplasm is 80% of the membrane and is usually colorless. The constituents of the structure were the disordered colloidal solution and the mass of the gel.
Sequence factors: The bacteria were used for alteration of the conversion of the genes and traits. This is used for the determination of the process in Fig. 1.
The sequence used three identifiers namely G, U, C and A characterization of constituents for the cytoplasm.
Spectrum of microorganisms: The cell suspensions contained quantitative information such as the number, size, structure, composition and interior of particles. The derivation of data resulted from transmission measurements of prokaryotic microorganisms.
Glycan array technologies: Glycosylation interactions were used for the characterization of the biology of glycoconjugates. The Chlamydomonas synthesis was studied in the experiment.
|
Phylogenetic method: This was used for the living organism for its regularity of Sec constituents for complete genome sequences in bacteria and plant divergences of eukaryotes. This had different characteristic rates.
Laboratory arrays were for the identification of the presence and functions of exoenzymes. These are processed through the Golgi apparatus and released into the cells.
RESULTS AND DISCUSSION
Hyphae were semiflexible 7 nm in diameter in the culture. Minus 5’ monomer concentration was approximately six times higher than the additional 3’ of 0.6 μM and 0.1 Μm. It was found cytosol a portion of the cytoplasm containment within membrane organelles. This made up 70% of cell volume and a mixture of hyphae dissolution of cells in Table 1.
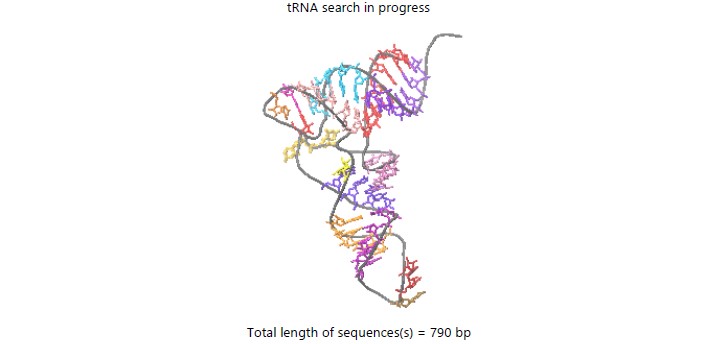
From computational data, it was revealed the cytoplasm was a dense solid mass of cytosol. It contained 32.1 gels and as high as 67.9%. Although this had an osmotic imbalance. The base number at the ends was equal to 790 in Fig. 2.
This suggested the cell surround was not permeable for diffusion process occurrences. Therefore, most of its constituents could only be revealed with the transmission spectroscopy method in Fig. 3.
The genome features affected phylogenetics for 155 diverse species of 7000 high-quality cells. The habitat flexibility was an approximate combination of particular genome sizes.
The bacteria were detected in the Hyphae with the G identifier, fungi with the T indicator and algae with the same for the cell. This when compared to the literature showed the asymmetry in cell division at high density. Therefore in references, the bacteria interact with the cell preferentially retained the template-DNA strands, reducing the spread of mutant cells1.
The inclusion of particles of insoluble substances in the cytosol. The recent measurement used spectrum microscopy indication of the cytoplasm2. This when compared to the literature had challenges in assigning molecular and cellular functions to thousands of newly predicted gene products. It was understood for events in molecular detail, the proteome must be analyzed3.

|
|
| Table 1: | Eukaryotic base numbers for the strands of cytoplasm | |||
| Search mode | Eukaryotic |
| Searching with | tRNAscan+EufindtRNA->Cove |
| Isotype-specific model scan | No |
| Covariance model | models/TRNA2-euk.cm |
| tRNAscan parameters | |
| EufindtRNA parameters | Relaxed (Int Cutoff= -32.10) |
| First-pass stats | |
| Sequences read | 1 |
| Seqs w/at least 1 hit | 0 |
| Bases read | 790 (x2 for both strands) |
| Bases in tRNAs | 0 |
| tRNAs predicted | 0 |
| Av. tRNA length | 0 |
| Script CPU time | 0.00 sec |
| Scan CPU time | 0.00 sec |
| Scan speed | 1580.0 Kbp sec–1 |
The membrane divided the cell from the surrounding environment4. This was semi-permeable and selectively for either a substance passage or a lower or none in the cell. This compared to the reference was a model for the quantitative interpretation of spectral patterns resulting from transmission measurements of prokaryotic microorganism suspensions. It was shown that spectroscopy techniques coupled with effective interpretation models apply to a wide range of cell types found in diverse environments5.
In bacteria and fungi, exoenzymes had importance for the allowance of organisms’ effective interaction in the environment Maistrenko et al.6. Some pathogenetic species used exoenzymes virulence factors for enablement of the diversity of virion. when compared to phages, these were essential for understanding microbial systems and their exploitation. It was shown the roles of phages in different host systems and how modeling, microscopy, isolation, genomic and metagenomic approaches have combined to provide insights into this small but vital constituents7.
From a preliminary search, there existed three species of bacteria and fungi in the cytoplasm. These were Bsp68I (bacteria) at base numbers 161 and 401, EcoRV at numbers 401 and 721 (fungi) and the presence of a eukaryote dissimilar from the prokaryotes at bases 721 of two algae HincII. This alga was not permeable by the cell exterior membrane8. Therefore was not considered present in the cytoplasm. But the osmotic imbalance was at the neighboring membrane.
The implication was image analysis could be used for the detection of processes in the Hyphae, particularly with the G identifier, fungi with the T indicator and algae with the same for cell1.
Research indicates new regions of exploration in the membrane divided the cell from the surrounding environment with hyphae4. This had to be semi-permeable and selectively for either a substance passage or a lower or none in the cell5. In bacteria and fungi, exoenzymes had importance for the allowance of organisms’ effective interaction in the environment6. Research should search existed in three species of bacteria and fungi in the cytoplasm7. This alga was not permeable by the cell exterior membrane8.
The implication was the mutation rate with complete genome sequences had spectra of microorganisms and cell suspensions moved by the actin network beneath the membrane within eukaryotic cells.
The application was exoenzyme had remarkable spatiotemporal organization inhabited by processes and absorption for the capability of bacteria and archaea diversity and important biological activities.
The recommendation was to study exoenzyme an extracellular enzyme secretion by cells and functions outside of the organism.
The limitation was this indicated the evolution rate at a high-density decrease of diversity for the non-division of pathogenic cells in the membrane of plants.
CONCLUSION
The cytosol identification was within prokaryotic genome sequences. The studies of cytoplasm synthesis in 93107 prokaryotic genomes characterization. BacStalk detected the divided regions of cells namely cell body, stalk and bud for complex bacteria. Cells were interaction links to the phylogenetics and processes. The quantitative identification of microorganisms such as size and mass concentration were elements of the organism. This indicated the evolution rate at a high-density decrease of diversity for the non-division of pathogenic cells in the membrane of plants.
SIGNIFICANCE STATEMENT
Spectra of microbes and cells travelling through an actin network beneath the membrane within eukaryotic cells are revealed by the mutation rate in full genome sequences. Exoenzymes possess a remarkable spatiotemporal organization that contains processes, absorbs the diversity of bacteria and archaea and supports crucial biological functions. Prokaryotes have a spatiotemporal organization of cellular functions. Enzymes frequently break down large molecules, allowing the constituents to flow through the cell membrane and enter the cell. Membrane organelles contain a component of the cytoplasm, known as the cytosol. Most constituents can only be revealed with the diffusion process since the cell surround is not permeable to diffusion process events.
ACKNOWLEDGMENT
The author thanks Association for Molecular Pathology for the participation in research studies and literature used for the article.
REFERENCES
- Aanen, D.K. and A.J.M. Debets, 2019. Mutation-rate plasticity and the germline of unicellular organisms. Proc. R. Soc. B, 286: 1902.
- Aebersold, R. and B.F. Cravatt, 2002. Proteomics-advances, applications and the challenges that remain. Trends Biotechnol., 20: S1-S2.
- Alley, W.R., B.F. Mann and M.V. Novotny, 2013. High-sensitivity analytical approaches for the structural characterization of glycoproteins. Chem. Rev., 113: 2668-2732.
- Clokie, M.R., A.D. Millard, A.V. Letarov and S. Heaphy, 2011. Phages in nature. Bacteriophage, 1: 31-45.
- Hartmann, R., M.C.F. van Teeseling, M. Thanbichler and K. Drescher, 2020. BacStalk: A comprehensive and interactive image analysis software tool for bacterial cell biology. Mol. Microbiol., 114: 140-150.
- Maistrenko, O.M., D.R. Mende, M. Luetge, F. Hildebrand and T.S.B. Schmidt et al., 2020. Disentangling the impact of environmental and phylogenetic constraints on prokaryotic within-species diversity. ISME J., 14: 1247-1259.
- Sanchez, I., R. Hernandez-Guerrero, P.E. Mendez-Monroy, M.A. Martinez-Nunez, J.A. Ibarra and E. Perez-Rueda, 2020. Evaluation of the abundance of DNA-binding transcription factors in prokaryotes. Genes, 11: 52.
- Skinnider, M.A., C.W. Johnston, N.J. Merwin, C.A. Dejong and N.A. Magarvey, 2018. Global analysis of prokaryotic tRNA-derived cyclodipeptide biosynthesis. BMC Genomics, 19: 45.
How to Cite this paper?
APA-7 Style
Ubani,
S.I. (2023). Constituents of Prokaryotic Organisms for Production of Extracellular Enzymes in the Development of Immunological Process. Asian Journal of Biological Sciences, 16(3), 234-239. https://doi.org/10.3923/ajbs.2023.234.239
ACS Style
Ubani,
S.I. Constituents of Prokaryotic Organisms for Production of Extracellular Enzymes in the Development of Immunological Process. Asian J. Biol. Sci 2023, 16, 234-239. https://doi.org/10.3923/ajbs.2023.234.239
AMA Style
Ubani
SI. Constituents of Prokaryotic Organisms for Production of Extracellular Enzymes in the Development of Immunological Process. Asian Journal of Biological Sciences. 2023; 16(3): 234-239. https://doi.org/10.3923/ajbs.2023.234.239
Chicago/Turabian Style
Ubani, Solomon, Ifeanyichukwu.
2023. "Constituents of Prokaryotic Organisms for Production of Extracellular Enzymes in the Development of Immunological Process" Asian Journal of Biological Sciences 16, no. 3: 234-239. https://doi.org/10.3923/ajbs.2023.234.239

This work is licensed under a Creative Commons Attribution 4.0 International License.



