Inhibition of Quorum Sensing Regulated Protein of E. coli using Food-based Biomolecules
| Received 18 Apr, 2022 |
Accepted 15 Jun, 2022 |
Published 01 Jul, 2022 |
Background and Objective: E. coli is one of the most important pathogens that cause nosocomial infections. As most microorganisms are becoming resistant to various drugs, the need for new Therapeutics is increasing day by day. Older methods mainly focused on killing the bacteria, which again makes them more resistant to new drugs. In this study, instead of killing the bacteria, the quorum sensing mechanism of the bacteria is interrupted using food biomolecules, thereby preventing the virulence activities of the bacteria. Materials and Methods: Bioactive compounds were selected based on their QQ activities and were subjected to molecular docking using AutoDock. The regulatory protein of E. coli (SdiA) was used as the target. After docking, the biomolecules with high docking scores were selected and carried out for in vitro analysis. The study was carried out in SLIMS and Divine Mother College, Pondicherry. Results: Out of 24 biomolecules, Epigallocatechin-3-gallate, naringin and Mangiferrin showed high docking scores against the protein SdiA. Epigallocatechin-3-gallate and naringin were used for in vitro studies and showed high anti-quorum-sensing activity in E. coli. They showed high inhibition of EPS and Biofilm formation in E. coli. Conclusion: To understand the QQ activity of the biomolecules and their mode of action further molecular interaction studies are needed. They can be used to develop efficient drugs.
| Copyright © 2022 Sundar et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Quorum Sensing (QS) is the mechanism of communication of bacteria within its cells. QS is achieved with the help of chemical signal molecules. When the bacterial population increases, Autoinducers (AIs) are accumulated in the environment and they help the bacteria to track changes in their cell count and help in maintaining gene expression. The control of many processes in bacteria like bioluminescence, antibiotic production, sporulation, competence, virulence activity and biofilm formation by Quorum sensing1-3. Nosocomial infections, often known as Healthcare-Associated Infections (HAI), are infections acquired while receiving health care that was not present at the time of admission. Nosocomial infections are the most common adverse event that compromises the safety of patients in health care. They can cause severe illness, demise and financial hardship for persons, their families and health care systems. The growth of multidrug-resistant microbes is another issue associated with nosocomial infections. In the United States, HAI affects 3.2% of all hospitalized patients, 6.5% in the European Union/European Economic Area and the global frequency is likely substantially greater4-6. E. coli is a Gram-negative, facultative anaerobic bacteria, which is a major causative organism for Gram-negative sepsis and endotoxin-induced shock. Other common nosocomial infections caused by this organism include infections of the urinary tract and wounds, pneumonia in immunocompromised hospitalized patients, meningitis in newborns and E. coli-associated diarrheal illness or gastroenteritis. Endotoxin, capsule, antigenic phase variation, sequestration of growth factors, serum killing resistance and antibiotic resistance are some of the virulence factors shared by E. coli and the Enterobacteriaceae family. However, some disease-causing strains, such as those that cause UTIs and gastroenteritis, have specialized virulence factors. Worldwide, Multidrug-Resistant (MDR) infections are a major cause of mortality; nearly 700,000 people die due to this each year and it is estimated that by 2050, there would be 10 million deaths each year7. This is due to the widespread use of existing antimicrobials in humans, animals and plants, which has resulted in the development of Multidrug-Resistant (MDR) harmful bacteria8. Antibiotics act upon the bacteria by interrupting their important processes. This leads to growth detention in bacteria which causes selective pressure in them making more MDR strains9.
LuxI/R type QS systems are used by Gram-negative bacteria and contain LuxI-type autoinducer synthases, which produce AHL (Acyl-Homoserine Lactone) molecules as Auto Inducers (AIs) and related LuxR-type receptor proteins. The transcriptional regulatory proteins LuxR-type receptors interact with AIs to mediate QS pathways. LuxR-type proteins such as LuxR, LasR and TraR are highly unstable without AIs and AI concentrations will be correspondingly low at low cell density10. When the cell density rises, the concentration of AI rises with it. These accumulating AHLs bind with the LuxR receptor, stabilizing the LuxR-AHL complex. The LuxR-AHL complex then attaches to the relevant promoter region, causing downstream genes to be expressed11. E. coli have a LuxR homologue, SdiA12, but do not have a luxI gene and do not produce AHLs12,13.
Current studies observed the evolution of multidrug resistance in bacteria and therefore the necessity of a novel antimicrobial approach that can disturb the bacterial communication by Quorum Sensing Inhibitors (QSI). According to studies, inhibiting the quorum signalling pathway can promote immune system function and contribute to the elimination of pathogenic microbes. The naturally occurring quorum-quenching mechanisms that efficiently inhibit microbial quorum sensing has been found in prokaryotes as well as eukaryotes. They act by hindering the key steps of quorum sensing, such as signal generation, signal accumulation, or signal reception14. The value of various food components in reducing various diseases, as well as the role of different probiotic content in dairy products in improving and avoiding gastrointestinal and other disorders, have all been proven in various studies. The importance and health benefits of personalized diets according to the risk group of an individual are reassured in many studies. The presence of active quorum sensing Inhibitors components in various food sources was relieved in many pieces of research. It was found that several crude extracts from food products help in the fast clearing of microorganisms. A study was conducted by Zimmer et al.15, where they found out blueberry fruit extracts showed marked activity against S. epidermidis biofilm without obstructing bacterial growth.
Studying food-derived components that can control or prevent quorum sensing can be helpful for the well-being of people and thus there arises a necessity to explore this field. In the present study, edible food biomolecules were screened for their quorum sensing inhibiting activity against the SdiA protein of E. coli using computer software like virtual screening and molecular docking. These techniques help us to identify possible anti-QS molecules against the SdiA of E. coli. They could be used as a food-based drug against E. coli infections after being tested in vitro and in vivo. The current study aimed to assess the quorum sensing inhibition activities using edible food biomolecules by in silico and in vitro methods against nosocomial infection causing multidrug-resistant E. coli.
MATERIALS AND METHODS
Study area: The present study was carried out at Sri Lakshmi Narayana Institute of Medical Science (SLIMS), Pondicherry and Divine Mother College, Pondicherry from January to March, 2022.
Selection of bioactive compounds: To understand docking interactions between edible bioactive compounds and quorum sensing regulatory protein of E. Coli (SdiA), a molecular docking study was made using AutoDock tools. The list of bioactive compounds from different foods which were selected based on previous research studies were listed in Table 1.
Retrieval of 3D structures of target and food compounds
Protein Data Bank (PDB): The Protein Data Bank (PDB) is a database that stores three-dimensional structural data for complex biological entities including proteins and nucleic acids. The data, which is normally collected by X-ray crystallography, Nuclear Magnetic Resonance (NMR) crystallography, or frequently cryo-electron microscopy and put forward by biologists and biochemists globally, are freely available on the Internet through the websites of its member organizations. In this paper, the high-resolution three-dimensional structures of the target protein, SdiA (PDB ID: 2VAX) are from the PDB database.
PubChem: PubChem is an open chemistry database at the National Centre for Biotechnology Information (NCBI). In this study, the PubChem database to download structures of 24 food biomolecules was used. The structures were downloaded in SDF format, which can be converted to PDB format using Pymol for molecular docking.
| Table 1: | Food source and bioactive compounds | |||
| Food sources | Bioactive compounds |
| Milk | Bovine Lactoferrin |
| Green tea | (-)-epicatechin ( EC) |
| (-)-epicatechin-3-gallate (ECG) | |
| (-)-epigallocatechin (EGC) | |
| (-)-epigallocatechin-3-gallate(EGCG) | |
| Zingiberofficinale | 6-gingerol |
| 8-gingerol | |
| 10-gingerol | |
| Curcuma amada | Cinnamic acid |
| Ferulic acid | |
| P-coumaric acid | |
| Protocatechic acid | |
| Gentisic acid | |
| Gallic acid | |
| Orange | Naringin |
| Neohesperidin | |
| Hesperidin | |
| Grapes | Resveratrol |
| Lemon | D-Limonene |
| Limonexic acid | |
| Mango | Mangiferin ( 2-β-D-glucopyranosyl-1,3,6,7-tetrahydroxy-9H-Xanthen-9-one) |
| Pumpkin, chilli, eggplant | Zeaxanthin |
| Fish | Docosahexaenoic acid |
| Eicosapentaenoic acid | |
| Cauliflower | Quercetin |
Molecular docking of bioactive compounds to target by AutoDock: The interaction of molecules underpins the majority of biological processes. These molecules could be proteins, enzymes, or medicines and the optimal result comes from a good interaction between them. Docking is the software’s prediction of these interactions. It aids in predicting the effectiveness of a protein’s interaction with our biomolecule. Due to the emerging needs for structural molecular biology and drug discovery based on structure, molecular docking has developed during the last three decades. As the availability and power of computers have increased nowadays, there is no difficulty in accessing small molecules and protein databases, which thereby facilitates the development of molecular docking16-19.
MGLTools is software for 3D visualization and analysis of molecular structures. The software includes three main applications: AutoDock Tools (ADT), Python Molecular Viewer (PMV) and Vision. AutoDock (http://autodock.scripps.edu) was used to do molecular docking in this study. It is a software mainly used for protein-ligand docking. It is one of the simplest and most cited software applications for docking in the research community20.
In vitro studies for Quorum Sensing Inhibition (QSI) activity
Minimum Inhibitory Concentration (MIC) of Epigallocatechin-3-Gallate (EGCG) and Naringin against E. coli: As suggested by the Clinical and Laboratory Standards Institute, USA (2006), MIC of EGCG and naringin was determined against E. coli. Overnight bacterial culture was inoculated into 20 mL of Lysogeny Broth (LB) medium. Different concentrations of EGCG and naringin ranging from 0.1-100 mg were incubated at 37°C for 24 hrs. Before and after incubation, the absorbance of the medium was measured at a wavelength of 600 nm. The lowest concentration of EGCG and naringin showed inhibition of growth was noted as the MIC.
Reduction in exopolysaccharide production: E. coli was grown in LB broth at 37°C in an inert condition. To obtain crude EPS, cells adhered to the test tube walls were gathered at the early stationary phase by centrifugation at 8000 rpm for 30 min at a temperature of 4°C. The filtrate was then passed through a membrane of 0.22 mm and three volumes of cold ethanol were added and incubated at a temperature of 2°C overnight to precipitate the EPS. The precipitated EPS was gathered by centrifugation for 30 min at 8000 rpm and dissolved in 1 mL of distilled water. The phenol-sulfuric acid method was used to measure the EPS21. The EPS reduction by Epigallocatechin-3-Gallate (EGCG) and Naringin were studied at their sub-MIC levels.
Inhibition of E. coli biofilms by microtitre plate assay: Biofilm inhibition assay was performed by microtitre plate assay. One% of overnight E. coli culture was inoculated to LB broth with and without Epigallocatechin-3-Gallate (EGCG) and Naringinin microtitre plates and incubated at 37°C. Planktonic E. coli cells were removed after incubation by carefully rinsing with sterile deionised water and surface adherent biofilms were stained for 10 min with 0.2% crystal violet. Crystal violet in biofilms was solubilized with 95% ethanol after the excess stain was drained. The crystal violet intensity at OD650 nm was measured with a microtitre plate reader to determine the biomass of s biofilms22.
RESULTS
Twenty-four bioactive compounds from different food sources were collected from the PubChem database and are used for molecular docking studies using AutoDock tools. The crystallographic structures of SdiA protein with PDB ID: 2VAX are used as the target. The molecular docking results and interactions between the edible food biomolecules and SdiA protein were collected.
The findings of in silico molecular docking studies revealed that phytochemicals found in natural resources may have a regulatory role. According to the molecular docking results, 10 food components, including
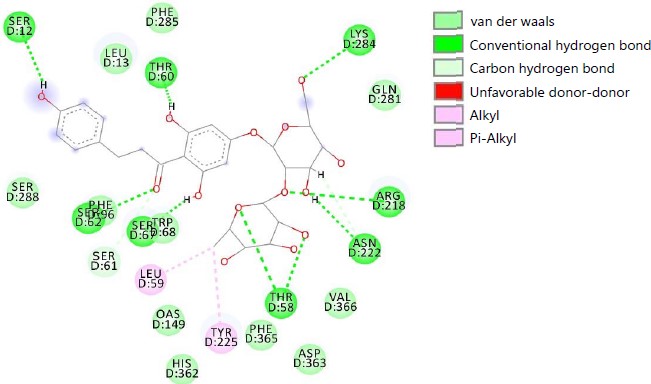 |
Fig. 1: 2D interaction diagram between SdiA and Epigallocatechin-3-gallate |
| Table 2: | Flexible docking of SdiA (PDB: 2VAX) with top ranked bioactive compounds | |||
| Compound ID | Compound names |
Ligand efficiency (kcal/mol) |
Dock score (kcal/mol) |
Binding energy (kcal/mol) |
| 102025303 | Epigallocatechin-3-gallate |
-0.234 |
-13.104 |
-76.111 |
| 9894584 | Naringin |
-0.267 |
-10.964 |
-65.074 |
| 5281647 | Mangiferrin |
-0.339 |
-10.156 |
-50.252 |
| 12795889 | Epicatechin-3-gallate |
-0.186 |
-9.857 |
-58.314 |
| 30231 | Neohesperidin |
-0.219 |
-9.405 |
-60.478 |
| 10621 | Hesperidin |
-0.216 |
-9.301 |
-55.073 |
| 65064 | Epigallocatechin |
-0.269 |
-8.882 |
-58.112 |
| 107905 | Epicatechin |
-0.261 |
-8.36 |
-55.321 |
| 370 | Gallic acid |
-0.6 |
-7.198 |
-28.675 |
| 5280343 | Quercetin |
-0.323 |
-7.095 |
-34.268 |
Epigallocatechin-3-gallate, Naringin, Mangiferrin, Epicatechin-3-gallate, Neohesperidin, Hesperidin, Epigallocatechin, Epicatechin, gallic acid and quercetin have high docking scores compared to the rest of the food biomolecules. The food biomolecules with the highest docking scores were listed in Table 2. The table indicates the compound ID, compound name, ligand efficiency, dock score and binding energy. The binding energy per atom of a ligand towards its binding partner, (receptor/enzyme) is calculated by ligand efficiency. It is measured in kcal/mol. Dock score is a scoring function used to predict the binding affinity between two molecules after molecular docking. It represents the change in potential energy during the
interaction between protein and ligand. So when the score is very negative, then the binding is strong and when the score is less negative or positive, then the binding is weak or non-existing. The docking score is measured in kcal/mol. In the table, Epigallocatechin-3-gallate and naringin have the highest dock score i.e., -13.104 and -10.964 respectively, when compared to other molecules. Therefore, it was selected as the top-ranked molecules and was subjected to in vitro analysis. Binding energy is the energy released by the protein-ligand interaction. It is measured in kcal/mol. A negative binding energy means, the ligand was bound spontaneously without consuming energy. Positive binding energy means the binding is energy consuming and it will occur only if the required energy is available. In the table, a high negative binding energy was given by Epigallocatechin-3-gallate and naringin ( -76.111 and -65.074, respectively).
Fig. 1 represented the 2D interaction between SdiA and Epigallocatechin-3-Gallate (EGCG) and the interaction of SdiA and naringin was represented in Fig. 2. Ribbon representation of SdiA protein docked with selected molecules was shown in Fig. 3.
 |
Fig. 3: Ribbon representation of SdiA protein docked with selected molecules |
The best two compounds EGCG and naringin were carried forwards for the in vitro studies based on the in silico results. Minimum Inhibitory Concentration (MIC) of EGCG and naringin was studied against E. coli and showed 400 μg mL–1 and 20 mg mL–1, respectively.
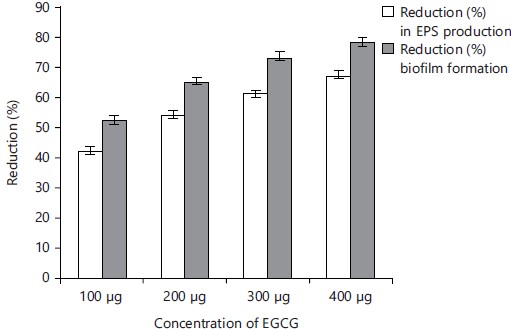
Inhibition of EPS production and biofilm formation by EGCG at sub-MIC levels were represented in Fig. 4. The X-axis represents the concentration of EGCG in μg and the Y-axis represents the % reduction of EPS and biofilm production by EGCG against E. coli. The blue bar represents %age reduction in EPS production and the orange bar represents a reduction in biofilm production by EGCG.
Concentration-dependent reductions in EPS production and E. coli biofilm formation of naringin were shown in Fig. 5. The X-axis represents the concentration of naringin in mg and the Y-axis represents a reduction of EPS and biofilm production in age (%) by naringin against E. coli. The blue bar represents age (%) reduction in EPS production and the orange bar represents a reduction in biofilm production by naringin.
 |
Fig. 5: Inhibition of EPS production and biofilm formation in E. coli by naringin |
EGCG inhibited a maximum of up to 67% EPS production in E. coli and naringin showed a 61% decrease in EPS production. Inhibition of biofilm formation in E. coli was 78% for EGCG and 72% for naringin.
DISCUSSION
In the present study, 10 out of 24 biomolecules showed high docking scores against the regulatory protein of E. coli. The best 3 biomolecules were Epigallocatechin-3-gallate, naringin and Mangiferin. From in vitro studies, Epigallocatechin-3-gallate and naringin showed inhibition of EPS and biofilm formation in the bacteria. Minimum Inhibitory Concentration (MIC) of EGCG and naringin was studied against E. coli and showed 400 and 20 mg mL–1, respectively.
Many plant-derived natural compounds have been identified as QSIs, although their specificity for LasR or other quorum regulators has been little explored. Grapefruit furocoumarins suppressed biofilm production by bacteria such as E. coli O157:H7, Salmonella typhimurium and Pseudomonas aeruginosa, as well as Auto Inducer-1 and Auto Inducer-2 activity in V. harveyi reporter, strains BB886 and BB170. Purified furocoumarins like dihydroxybergamottin and bergamottin inhibited Auto Inducers(AIs) in the 94.6-97.7% range. Grape juice, on the other hand, inhibited AI-1 and AI-2 by 47–62 and 16.8–27.5%, respectively. Low concentrations of furocoumarins in grapefruit juice were blamed for the weaker
inhibitory effects23. The QSI actions of flavonoids such naringenin, kaempferol, quercetin and apigenein were investigated. In V. harveyi BB886 and MM32, all of these flavonoids blocked HAI-1 or AI-2-mediated bioluminescence. V. harveyi BB120 and E. coli O157:H7 were reported to suppress biofilm formation by quercetin and naringenin24. Quorum quenching effects have also been reported in extracts from Combretum albiflorum, Laurus nobilis and Sonchus oleraceus leaves, flowers, fruit and bark25,26. Quorum sensing inhibition has been associated with garlic27,28, medicinal herbs29 and dietary components30-32. Catechins, found in tea and other herbal plants, prevent conjugative R plasmid transfer in E. coli. Grapefruit juice (furocoumarins) inhibited the production of E. coli biofilms23.
In this study, the QS regulatory protein of E. coli, (SdiA) was subjected to molecular docking with twenty-four bioactive compounds from various food sources to find their quorum sensing inhibitory properties. Structures of biomolecules were gathered from the PubChem database and the solution structure of the regulatory protein, SdiA, with PDB ID:2VAX is used as the target. The molecular docking scores of the top-ranked 10 compounds were listed in Table 2, ranking has been based on the "dock score (highest negative to a lowest negative value)". Quorum sensing enables E. coli in regulating their virulence activities and helps in cell division. It is easier to prevent virulence activities than to interrupt them once already established. Antibiotic resistance is a developing feature in microorganisms, therefore approaches that aid in the discovery of effective drugs against these pathogens is necessary. Using natural compounds against the target protein is a useful method for drug design. These can be done through in silico studies and will be useful to detect the molecular process as well as the public health threat of antimicrobial resistance.
In vitro studies also proved the quorum quenching activities of selected compounds EGCG and naringin. These compounds can serve as a Quorum Sensing Inhibition (QSI) based drug against E. coli infections spread through hospitals to overcome antimicrobial resistance.
CONCLUSION
Overall, the result of this in silico study to determine the anti-quorum sensing activity of twenty-four edible biomolecules from various food sources, against the regulatory protein of E. coli, SdiA, found that 10 molecules have high docking scores and can be used as quorum sensing inhibitors of E. coli. For in vitro and in vivo studies, the molecules that have a high docking score have to be selected. To understand the quorum quenching activities and the mode of action of the molecules in detail, further molecular interaction studies are needed. These molecules with quorum quenching activities can be used to develop new efficient drugs and can be used effectively along with the current antibiotics.
SIGNIFICANCE STATEMENT
This study discovered that nosocomial infections caused by E. coli can be inhibited without killing the bacteria using antibiotics. Food biomolecules can be used instead of antibiotics, to interrupt the quorum sensing mechanism of the bacteria and thus stop the infection. This helps to prevent the development of multidrug-resistant bacteria. This study will help the researchers to uncover the critical areas of multidrug- resistant bacteria and nosocomial infections that many researchers were not able to explore. Thus, a new theory on the Inhibition of quorum sensing regulated protein of E. coli using food-based biomolecules arrived.
REFERENCES
- Novick, R.P. and E. Geisinger, 2008. Quorum sensing in staphylococci. Annu. Rev. Genet., 42: 541-564.
- Ng, W.L. and B.L. Bassler, 2009. Bacterial quorum-sensing network architectures. Annu. Rev. Genet., 43: 197-222.
- Williams, P. and M. Cámara, 2009. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol., 12: 182-191.
- Magill, S.S., E. O’Leary, S.J. Janelle, D.L. Thompson and G. Dumyati et al., 2018. Changes in prevalence of health care-associated infections in U.S. hospitals. N. Engl. J. Med., 379: 1732-1744.
- Suetens, C., K. Latour, T. Kärki, E. Ricchizzi and P. Kinross et al., 2018. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: Results from two European point prevalence surveys, 2016 to 2017. Eurosurveillance.
- Allegranzi, B., S.B. Nejad, C. Combescure, W. Graafmans, H. Attar, L. Donaldson and D. Pittet, 2011. Burden of endemic health-care-associated infection in developing countries: Systematic review and meta-analysis. Lancet, 377: 228-241.
- Spaulding, C.N., R.D. Klein, H.L. Schreiber, J.W. Janetka and S.J. Hultgren, 2018. Precision antimicrobial therapeutics: The path of least resistance? npj Biofilms Microbiomes.
- Gupta, V. and L. Singhal, 2018. Antibiotics and Antimicrobial Resistance. In: Infectious Diseases and Your Health, Singh, P.P. (Ed.), Springer, Singapore, ISBN: 978-981-13-1577-0, pp: 215-224.
- Durão, P., R. Balbontín and I. Gordo, 2018. Evolutionary mechanisms shaping the maintenance of antibiotic resistance. Trends Microbiol., 26: 677-691.
- Smith, C., H. Song and L. You, 2008. Signal discrimination by differential regulation of protein stability in quorum sensing. J. Mol. Biol., 382: 1290-1297.
- Papenfort, K. and B.L. Bassler, 2016. Quorum sensing signal-response systems in gram-negative bacteria. Nat. Rev. Microbiol., 14: 576-588.
- Pacheco, T., A.É.I. Gomes, N.M.G. Siqueira, L. Assoni, M. Darrieux, H. Venter and L.F.C. Ferraz, 2021. SdiA, a quorum-sensing regulator, suppresses fimbriae expression, biofilm formation, and quorum-sensing signaling molecules production in Klebsiella pneumoniae. Front. Microbiol.
- Walters, M. and V. Sperandio, 2006. Quorum sensing in Escherichia coli and Salmonella. Int. J. Med. Microbiol., 296: 125-131.
- Dong, Y.H., L.H. Wang and L.H. Zhang, 2007. Quorum-quenching microbial infections: Mechanisms and implications. Philos. Trans. R. Soc. B: Biol. Sci., 362: 1201-1211.
- Zimmer, K.R., C.H. Blum-Silva, A.L.K. Souza, M. WulffSchuch and F.H. Reginatto et al., 2014. The antibiofilm effect of blueberry fruit cultivars against Staphylococcus epidermidis and Pseudomonas aeruginosa. J. Med. Food, 17: 324-331.
- Hendlich, M., 1998. Databases for protein-ligand complexes. Acta Crystallogr. Sect. D: Struct. Biol., D54: 1178-1182.
- Hu, L., M.L. Benson, R.D. Smith, M.G. Lerner and H.A. Carlson, 2005. Binding MOAD (mother of all databases). Proteins, 60: 333-340.
- Irwin, J.J. and B.K. Shoichet, 2005. ZINC-A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model., 45: 177-182.
- Berman, H.M., J. Westbrook, Z. Feng, G. Gilliland and T.N. Bhat et al., 2000. The protein data bank. Nucl. Acids Res., 28: 235-242.
- Morris, G.M., R. Huey and A.J. Olson, 2008. Using autodock for ligand-receptor docking. Curr. Protoc. Bioinf., 24: 8.14.1-8.14.40.
- Huston, A.L., B. Methe and J.W. Deming, 2004. Purification, characterization, and sequencing of an extracellular cold-active aminopeptidase produced by marine psychrophile Colwellia psychrerythraea strain 34H. Appl. Environ. Microbiol., 70: 3321-3328.
- Limsuwan, S. and S.P. Voravuthikunchai, 2008. Boesenbergia pandurata (Roxb.) Schltr., Eleutherine americana Merr. and Rhodomyrtus tomentosa (Aiton) Hassk. as antibiofilm producing and antiquorum sensing in Streptococcus pyogenes. FEMS Immunol. Med. Microbiol., 53: 429-436.
- Girennavar, B., M.L. Cepeda, K.A. Soni, A. Vikram and P. Jesudhasan et al., 2008. Grapefruit juice and its furocoumarins inhibits autoinducer signaling and biofilm formation in bacteria. Int. J. Food Microbiol., 125: 204-208.
- Vikram, A., G.K. Jayaprakasha, P.R. Jesudhasan, S.D. Pillai and B.S. Patil, 2010. Suppression of bacterial cell-cell signalling, biofilm formation and type III secretion system by citrus flavonoids. J. Appl. Microbiol., 109: 515-527.
- Al-Hussaini, R. and A.M. Mahasneh, 2009. Microbial growth and quorum sensing antagonist activities of herbal plants extracts. Molecules, 14: 3425-3435.
- Schaefer, A.L., E.P. Greenberg, C.M. Oliver, Y. Oda and J.J. Huang et al., 2008. A new class of homoserine lactone quorum-sensing signals. Nature, 454: 595-599.
- Bjarnsholt, T., P.Ø. Jensen, T.B. Rasmussen, L. Christophersen and H. Calum et al., 2005. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infectin. Microbiology, 151: 3873-3880.
- Rasmussen, T.B., T. Bjarnsholt, M.E. Skindersoe, M. Hentzer and P. Kristoffersen et al., 2005. Screening for Quorum-Sensing Inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol., 187: 1799-1814.
- Adonizio, A., K.F. Kong and K. Mathee, 2008. Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa by South Florida plant extracts. Antimicrob. Agents Chemother., 52: 198-203.
- Choo, J.H., Y. Rukayadi and J.K. Hwang, 2006. Inhibition of bacterial quorum sensing by vanilla extract. Lett. Appl. Microbiol., 42: 637-641.
- Vattem, D.A., K. Mihalik, S.H. Crixell and R.J.C. McLean, 2007. Dietary phytochemicals as quorum sensing inhibitors. Fitoterapia, 78: 302-310.
- Zhu, H. and S.J. Sun, 2008. Inhibition of bacterial quorum sensing-regulated behaviors by Tremella fuciformis extract. Curr. Microbiol., 57: 418-422.
How to Cite this paper?
APA-7 Style
Sundar,
K., ,
S.J., Guru,
K.K., Jayalakshmi,
G. (2022). Inhibition of Quorum Sensing Regulated Protein of E. coli using Food-based Biomolecules. Asian Journal of Biological Sciences, 15(4), 249-258. https://doi.org/10.3923/ajbs.2022.249.258
ACS Style
Sundar,
K.; ,
S.J.; Guru,
K.K.; Jayalakshmi,
G. Inhibition of Quorum Sensing Regulated Protein of E. coli using Food-based Biomolecules. Asian J. Biol. Sci 2022, 15, 249-258. https://doi.org/10.3923/ajbs.2022.249.258
AMA Style
Sundar
K,
SJ, Guru
KK, Jayalakshmi
G. Inhibition of Quorum Sensing Regulated Protein of E. coli using Food-based Biomolecules. Asian Journal of Biological Sciences. 2022; 15(4): 249-258. https://doi.org/10.3923/ajbs.2022.249.258
Chicago/Turabian Style
Sundar, Kothandapani, Sherin Joseph , K. Kumara Guru, and G. Jayalakshmi.
2022. "Inhibition of Quorum Sensing Regulated Protein of E. coli using Food-based Biomolecules" Asian Journal of Biological Sciences 15, no. 4: 249-258. https://doi.org/10.3923/ajbs.2022.249.258

This work is licensed under a Creative Commons Attribution 4.0 International License.