Purification and Characterization of an Alkaline Protease from Aspergillus fumigatus Cas1 Isolated from Industrial Polluted Area
| Received 07 Jan, 2022 |
Accepted 10 Oct, 2022 |
Published 31 Mar, 2023 |
Background and Objective: Protease was an important industrial enzyme with high demand. Finding an efficient alkaline protease was a difficult task for the researchers. Alkaline protease enzyme from fungus Aspergillus fumigatus CAS1, isolated from SIPCOT (State Industries Promotions Corporations of Tamil Nadu) near Cuddalore was the aim of this study. Materials and Methods: The fungal strain was identified by using 18S rRNA molecular identification. Different carbon sources such as skim milk, glucose, sucrose and seaweed were used for the production of enzymes. Results: Among the carbon sources, skim milk (820 U mL‾1) showed maximum enzyme production, followed by seaweed (Sargassum tenerrimum) (650 U mL‾1). In nitrogen sources (casein, yeast extract, soybean meal, peptone and fish wastes) maximum enzyme production in casein (685 U mL‾1), followed by fish wastes (Sardinella longiceps). The optimum pH is 9 (380 U mL‾1) and temperature 55 (420 U mL‾1) for effective protease production. In DEAE cellulose enzyme purification 17.29% recovery was achieved with 4.3 fold purification. The molecular mass was estimated to be 66KDa on SDS-PAGE. Conclusion: The enzyme alkaline protease obtained from the isolate A. fumigatus CAS1 showed astonishing results in the wash performance analysis and dehairing.
| Copyright © 2023 Sugesh et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Total enzyme sales protease constitute 60% of all other industrial enzymes1,2. Thermostable and alkaline proteases have a wide range of single applications in food processing, detergent making, beverage production, animal nutrition, leather process, paper, pulp, textile and pharmaceutical industries and also in wastewater treatment3,4. A ‘N’ number of microbes (bacteria, fungi, yeast, moulds, etc.) are producing an extra and intracellular protease, which fungal proteases are unique. e.g., Aspergillus fumigatus TKU003, A. niger, Penicillium roqueforti, Trichoderma harzianum and Cephalosporium sp., KM3885-8. The demand for industrial proteolytic enzymes which have a unique nature like specificity and stability of pH, temperature, utilization of complex and synthetic nutrients, etc., stimulates the search for unique enzymes from new sources.
Proteases with high activity, stability, high temperature and high range of alkalinity were shown the extensive area of research in bioengineering and biotechnological application9-11. Thermostable proteases are advantageous as high production and temperature processing are capable of increasing reaction rates, solubility, products and also decreasing microbial contamination. The worldwide need for thermostable enzymes is two-thirds of all other proteases12. Some thermophilic fungal species are producing unique proteases such as Thermoascus aurantiacus13 and A. niger14. Any have only less number of reports are available in the production of thermophilic proteases from fungi. Sivasanker et al.15 had reported L-asparaginase from a natural substrate such as mangrove leaves, seaweed, bivalve extract, gastropod extract, among all the natural source mangrove leaf extract Avicennia marina produced a large amount of L-asparaginase production than that of other sources.
To obtain industrially important biocatalyst has to screen new strains from nature. In this view, the present investigation was successfully isolated a highly potent protease producing strain A. fumigatus from SIPCOT (State Industries Promotion Corporation of Tamil Nadu) (Lat 11°42'23.15"N, 79°46'57.97"E) near Cuddalore, India. This strain was producing a substantial amount of protease in solid substrate culture. A first attempt by using seaweed for the production of proteases and also tried in fish wastes, different carbon and nitrogen source for the production of proteases.
MATERIALS AND METHODS
Study area: The study was carried out at CAS in marine biology, Annamalai University, Chidambaram, Tamil Nadu, India from March, 2015 to February, 2016. This study was analyzed from a state industrial promotion corporation in Tamil Nadu formerly known as SIPCOT. Many pharmaceutical and pesticide-based industries are running in SIPCOT. These industries release highly polluted toxic compounds to the environment. The soil and water samples were taken from this highly polluted environment because these environmental parameters facilitate the microbial population to produce unique secondary metabolites.
Initial screening: Sixteen strains were isolated at sediment samples from SIPCOT (Lat 11°42'23.15"N, 79°46'57.97"E) near Cuddalore. The isolated strains were screened using Czapeck Dox Broth (CDB) containing 1% casein for protease production. The CDB medium with 1% casein was prepared, adjusted to pH 7, dispensed into 50 mL each in 250 mL Erlenmeyer flasks and sterilized in an autoclave. After cooling the flask were inoculated with two mycelial discs (8 mm) of respective fungus and incubated at a rotary shaker (37°C, 180 RPM) for 7 days. The culture filtrates were separated by filtering through two-layered cheesecloth and centrifuged at 8000 RPM for 15 min. The mycelium free culture filtrate was prepared and used for the qualitative protease production assay.
Protease assay: To determine protease production used water agar medium of Carrim et al.16 (1.8 g agar in 100 mL of distilled water) supplemented with 1% casein was poured into Petri dishes and after solidification, 8 mm diameter wells were made using a cork borer. The 100 mL of culture filtrate was inoculated in a well and the same volume of un-inoculated medium was poured in a separate well as a control. After 24 hrs of incubation, the enzyme activity was visualized as a clear zone addition of 1% mercuric chloride solution in 1N HCl. Microorganisms showing clear zone around the colonies were marked as positive strains and it was sub-cultured. The morphology and biochemical characteristic were studied. Further molecular characterization was done by the 18S rRNA gene sequencing method.
18S rRNA gene sequencing: Miki et al.17 method was followed for the extraction of fungal DNA. In PCR amplification ITS1, ITS2, ITS3 and ITS4 were used18. Nucleotide sequencing was done with an automated DNA sequencing method followed by DNA hybridization19 with the use of universal primers. A BLAST search was done in the sequence to identify a similar sequence in the NCBI database. The 18S rRNA gene sequence accession number is JX283351.
Optimal parameters for protease production: Based on the results of the initial screening, the selected fungal strain A. fumigatus CAS1 was selected for parameter optimization. In the present study, the optimizing the fungal strain A. fumigatus CAS1 for the following parameters like temperature (25-55°C), pH (3.0-12), carbon sources (glucose, sucrose and skim milk), nitrogen sources (casein, yeast extract, peptone, Na2H2PO4 and soybean meal) which were expected to affect the production of protease.
Effect of fish wastes on protease production: The fish (S. longiceps) waste like viscera was collected from the local market Annan Kovil in Potonova. The fish wastes were washed with clear water and the meat was boiled at 50°C for 10 min. The water was removed and the meat was homogenized with mortar and pestle. The homogenized content was dried in shadow for 3-4 days. Then, dried content was ground for obtaining powder for optimizing protease production used as a nitrogen source. Different concentration (0.5-3 g) of fish wastes was used for the growth kinetic studies.
Effect of seaweed on protease production: A preliminary attempt has been made to use seaweed as a carbon source for the production medium for the protease enzyme. Seaweed (Sargassum tenerrimum) was freshly collected from the Mandabam study area and they were washed with water to remove salt content. Then seaweed was dried under shadow conditions for two to three weeks and powdered. The powdered content was at different concentrations (0.5-3 g) used for optimization studies.
Method of fermentation: Spores of A. fumigatus from 7 days old PDA slant were dislodged into 0.1% of tween-80 medium and it was used as an inoculum. The inoculums were adjusted using a hemocytometer for desired spore count20. For mass scale culture, 1 L of optimized media of protease production medium of Singh et al.21 (skim milk-12 g, peptone-9 g, KH2PO4-1 g, MgSO4.7H2O-0.2 g, Na2CO3-1 g, pH-9 and distilled water-1000 mL) was prepared in a 2 L conical flask. After sterilization of 2 mL of A. fumigatus spore suspension containing 1×105 spores, mL–1 was inoculated and incubated for 5 days in a rotary shaker at 200 RPM and 40°C. It was grown for five days after which the mycelium free supernatant was used for the purification.
Enzyme purification: The various steps of enzyme purification were carried out at 4°C. In an initial purification step, the supernatant containing the extracellular protease was treated with different saturation levels of solid ammonium sulfate (up to 80% saturation level), with continuous overnight stirring22. The precipitated protein was collected by centrifugation (10,000 RPM for 15 min) and dissolved in 0.1 M citrate phosphate buffer (pH 5). The enzyme solution was dialyzed in a dialysis membrane No. 150 (Himedia) against the same buffer for 48 hrs with several intermittent buffer changes. The partially purified protein was obtained and lyophilized into a powder. The lyophilized sample was dissolved in phosphate buffer (10 mL: pH 7) and loaded with DEAE cellulose column (GENEI) pre-equilibrated with 10 mL phosphate buffer (pH 7.0). The enzyme was eluted from the column using the linear gradient of NaCl (100-500 mm). Each fraction was quantitatively estimated for protease activity and protein content.
Protein estimation: The total protein content of the samples was determined according to the modified Lowery method described by Lucas and Tomazic-Jezic23, by using bovine serum albumin (BSA) as a standard. One unit of enzyme activity is defined as the amount of enzyme that liberates peptide fragments equivalent to 1 mg of BSA under the assay conditions24,45.
Polyacrylamide gel electrophoresis: Nowakowski25 method was used for the determination of the molecular mass of enzymes using known standard compounds.
Wash performance analysis: Different strains were used for wash performance studies such as Blood, ink and banana stains. White cloth 5×5 cm was stained with 25 μL of human blood, ink, banana stain and subjected to the following wash performance:
• |
Wash with tap water (negative control) at 28±2 |
• |
Wash with commercial detergents (positive control) at 28±2 |
• |
Wash with crude enzyme (50 mL) at 28±2 |
Dehairing: Dehairing of goatskin was performed using the dip method26. The two pieces of skin 4×4 (weighed around 2-4 g) of a goat was taken. One piece of goat hide was soaked on the crude enzyme-containing container (50 mL) and another hidden piece was taken as control which one was not treated with the crude enzyme. Both hides dehairing of goatskin was performed using the dip method26. The two pieces of skin 4×4 (weighed around 2-4 g) of a goat was taken. One piece of goat hide was soaked on the crude enzyme-containing container (50 mL) and another hidden piece was taken as control which one was not treated with the crude enzyme. Both hides were incubated at 45°C for 24-48 hrs on a rotary shaker at 120 RPM. After incubation both hides were analyzed for dehairing activity and also observed colour change in hides after removal of hair.
RESULTS AND DISCUSSION
A sum of sixteen fungal species was screened from sediment samples from SIPCOT near Cuddalore. They are divided into three categories based on the hydrolysis of casein, high producers (+++), medium producers (++), weakest producers (+) and negative producers (-). Out of sixteen isolates, only four strains were showed acceptable protease production. Among these, four isolates SS7 showed a larger zone of casein hydrolysis and it was selected for the optimization of protease production and characterization of protease. The isolate SS7 was identified as A. fumigatus based on morphological and physical characterization using standard mycological manuals27.
The isolate was subjected to 18S rRNA molecular identifications. The sequence results were compared with other sequences results obtained from NCBI-BLAST. The sequence showed 17% similar to other species of Aspergillus. In a phylogenetic analysis by a neighbour-joining method, the isolate SS7 showed homogeneity with distinct linkage with A. fumigators (Fig. 1). The most conserved region of the A. fumigatus CAS1 JX283351 was obtained through Seaview V4 software. It showed that A. fumigatus have 1-11 base pair differences and it has 0.009-0.024 (pi distances) pairwise differences with other species of Aspergillus. In overall observation of isolate SS7 (A. fumigatus CAS 1 JX283351) sequence differed from (1-11) excluded (alignment gaps) with intraspecies of Aspergillus sp. and it has <17% differences with other species of Aspergillus. Hajji et al.28 have isolated an effective protease producing strain A. clavatus from wastewater.
Effect of carbon source: The carbon sources, skim milk were showed predominant production and maximum production was achieved using a basal medium supplement of 3% (w/v) skim milk (820 U mL–1) (Fig. 2). In addition, A. fumigatus CAS1 produce protease when grown basal medium supplement with 3% (w/v) glucose (682 U mL–1) and 3% (w/v) sucrose (480 U mL–1). These results were somewhat different from earlier were reports studied in fungus A. fumigatus TKU00329 and Mucor miehei30.
Enhancing protease production in seaweed: Seaweed (S. tenerrimum) showed a predominant production of proteases and the maximum amount of production was achieved in basal medium 2% (w/v) seaweed (564 U mL–1) (Fig. 2). Seaweeds contain a high amount of minerals, vitamins and proteins and low content in lipid31,32. Seaweeds also are an essential source of fatty acid, they are eicosapentaenoic acid, C20:5ω333. Hence, these essential compounds may influence the growth of A. fumigatus and produce a high amount of protease enzyme.

|

|
Effect of nitrogen source: In addition to the organic nitrogen sources examined, casein showed the highest production rate protease in basal medium with a supplement of 3% (w/v) casein (940 U mL–1) (Fig. 3). When compared with casein a basal medium with a supplement of 2.5% (w/v) yeast extracts (320 U mL–1), soybean meal (360 U mL–1), peptone (345 U mL–1) and Na2H2PO4 (283.30 U mL–1). Similar results were also observed from fungal strains A. fumigatus TKU00329, A. clavatus34 and M. miehei30.
Enhancing the production of protease in fish wastes: All the organisms have their desired requirements for maximum enzyme production35. Therefore, medium optimization is essential for the production of enzymes at low cost and safe consumption. Generally, proteolytic microorganisms can grow only in media containing peptone, as a rich source of amino acids, peptides, proteins are becoming one of the most important constituents of culture media widely used for the production of a variety of metabolites, including enzymes22. On such aspects present study was tried with fish wastes as a nitrogen source for the production of protease and the fish wastes showed very high production rates (590 U mL–1) with the supplement of 3% (w/v) (Fig. 3).

|
Similarly, the shellfish wastes were used for the production of proteases36 and the fish processing wastes (viscera) were used for alkaline protease, fish sauce production37,38. Viscera of Catla catla were used for the production of protein hydrolysates39,40.
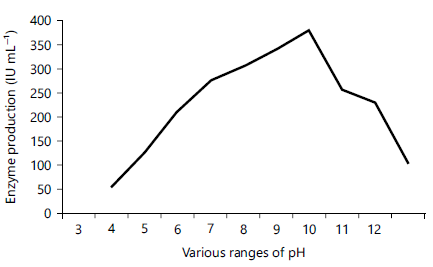
Effect of pH production of proteases: Among the physio-chemical parameters pH was an essential parameter that affects the production of enzymes41. To study the effect of medium pH on enzyme production in SMF, experiments were performed with medium different pH ranges between 3-12 and incubated for 5 days. The maximum production of protease was observed at pH 9 (590 U mL–1) and minimum production was occurring at pH 12 (105 U mL–1) (Fig. 4). Similar several findings showed pH 9 optimum for fungal proteases such as A. fumigatus42-44, A. parasiticus45, A. clavatus CCT275946 and A. fumigatus TKU00329.
Effect of temperature on production of protease: It is known that temperature is one of the most critical parameters that are controlled in any bioprocess47. The temperature range between 25-55°C was examined in the present study. The maximum production protease was observed in temperature 40°C (420 U mL–1) and the minimum was observed in temperature 55°C (150 U mL–1) (Fig. 5). The present investigation was supported by earlier works of Uyar and Baysal48. The temperature seemed to be higher compared to that of A. clavatus CCT275934 and A. fumigatus TKU00329 strain showed maximum protease activity between 37-42.8°C.
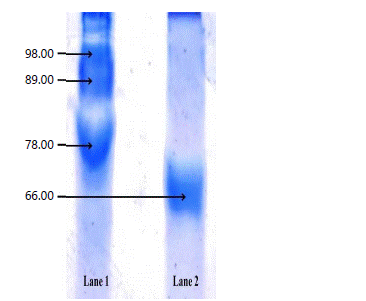
Enzyme purification: The protease activity of the crude enzyme extract was concentrated by using ammonium sulfate precipitation. So, 92.94% recovery of proteas with 2.8 fold purification was attained (Table 1). Following ammonium sulfate precipitation the enzyme sample was lyophilized. The partially purified lyophilized protease enzyme was made into a solution by dissolving in phosphate buffer (10 mm, pH 7) and this solution was applied to the DEAE cellulose column. Table 1 shows the purification profile of A. fumigatus. In the DEAE column, 17.79% were recovered with 4.3-fold purifications and 35.83 U mg–1 of a specific activity. A comparable result was reported in A. parasiticus showing that 90% recovery by 1.6 fold purifications. Likewise, Venugopal and Saramma49 have observed in Vibrio fluvialis with 95.5% recovery in 1.25 fold purification in 80% of ammonium sulfate precipitation. Purified protease from A. fumigatus showed 66 kDa on the SDS page and it’s suggested that the purified protein was homogenized (Fig. 6).
|

|
| Table 1: | Summary of purification procedure of protease from A. fumigatus CAS1 | |||
| Purification step (U) | Total activity (mg) |
Total protein (U mg–1) |
Specific activity |
Fold purification (%) |
Recovery |
| Crude extract (NH4)2SO4 | 16550 |
978.9 |
18.88 |
1 |
100 |
| Precipitate | 14300 |
488.4 |
23.14 |
2.8 |
92.94 |
| Dialyis | 9575 |
262.6 |
31.23 |
3.7 |
82.36 |
| DEAE-cellulose | 2750 |
87.3 |
35.83 |
4.3 |
17.79 |
Commercial application of protease enzyme
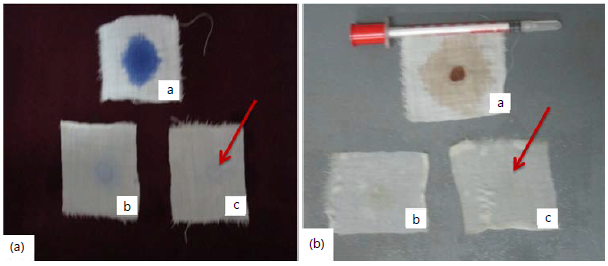
Washing performance: In the recent past, the detergent industry highly relies on microbial alkaline enzyme categories. Also, the detergent industries need the most dynamic microbial alkaline protease enzyme that withstand high temperature, pH and it necessarily possesses the presence of surfactants or oxidizing agent50. Similarly, the alkaline protease from the isolate SS7 (A. fumigatus CAS1) was shown extraordinary results. The partially purified enzyme removed both blood and ink stains more efficiently (Fig. 7a-b). But it showed some acceptable results on banana stain removal. Banik and Prakash51 have studied laundry detergent comparability of alkaline protease from Bacillus cereus. Rao et al.52 have reported characterization of thermo and detergent stable serine protease from Bacillus circulans and evaluation of eco-friendly applications. Savitha et al.53 has revealed the fungal protease, production and purification and comparability with laundry detergents and their wash performance.
|
|
|
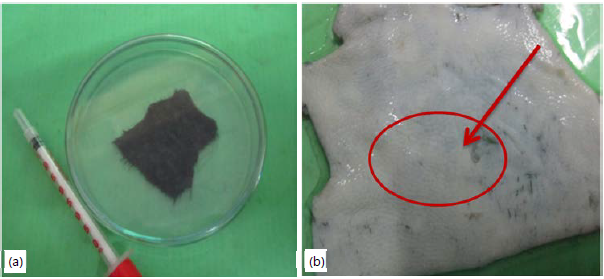
Dehairing: Mechanical plucking method was followed on dehairing assay. Easy removal of hairs was observed in enzyme treats hide and also not much colour change was observed. Whereas, in control no hair removal was detected (Fig. 8a-b). Sundararajan et al.54 stated that alkaline protease from Bacillus cereus VITSN04 has potential dehairing activity. Shrinivas and Naik55 have expressed highly stable alkaline protease from thermo-alkalophilic Bacillus halodurans JB 99 shown significant dehairing activities.
CONCLUSION
Based on the above observation, the present study was concluded that the isolate A. fumigatus CAS1 JX283351 was produced a high amount of enzyme production in different carbon and nitrogen sources. The enzyme alkaline protease obtained from the isolate A. fumigatus CAS1 showed astonishing results in the wash performance analysis and dehairing. This is the first report that we produced alkaline protease in seaweed (Sargassum tenerrimum) as a carbon source and fish wastes (S. longicepsis) as a nitrogen source.
SIGNIFICANCE STATEMENT
This study discovers efficient alkaline protease from A. fumigatus CAS1 JX283351 and it has shown astonishing results in wash performance analysis and dehairing activities. The A. fumigatus CAS1 JX283351 was producing alkaline protease enzymes from cheaper sources of fish wastes and seaweed. This study helped find novel industrial potent enzymes from highly complicated environments.
REFERENCES
- Gupta, R., Q. Beg and P. Lorenz, 2002. Bacterial alkaline proteases: Molecular approaches and industrial applications. Appl. Microbiol. Biotechnol., 59: 15-32.
- do Nascimento, W.C.A. and M.L.L. Martins, 2004. Production and properties of an extracellular protease from thermophilic Bacillus sp. Braz. J. Microbiol., 35: 91-96.
- Manavalan, T., A. Manavalan, S. Ramachandran and K. Heese, 2020. Identification of a novel thermostable alkaline protease from Bacillus megaterium-TK1 for the detergent and leather industry. Biology.
- Pastor, M.D., G.S. Lorda and A. Balatti, 2001. Protease obtention using Bacillus subtilis 3411 and amaranth seed meal medium at different aeration rates. Braz. J. Microbiol., 32: 6-9.
- Aalbæk, T., M. Reeslev, B. Jensen and S.H. Eriksen, 2002. Acid protease and formation of multiple forms of glucoamylase in batch and continuous cultures of Aspergillus niger. Enzyme Microb. Technol., 30: 410-415.
- Guo, Y., X. Li, W. Jia, F. Huang, Y. Liu and C. Zhang, 2021. Characterization of an intracellular aspartic protease (PsAPA) from Penicillium sp. XT7 and its application in collagen extraction. Food Chem.
- Dunaevsky, Y.E., T.N. Gruban, G.A. Beliakova and M.A. Belozersky, 2000. Enzymes secreted by filamentous fungi: Regulation of secretion and purification of an extracellular protease of Trichoderma harzianum Biochem. (Moscow), 65: 723-727.
- Lakshmi, B.K.M., D.M. Kumar and K.P.J. Hemalatha, 2018. Purification and characterization of alkaline protease with novel properties from Bacillus cereus strain S8. J. Genet. Eng. Biotechnol., 16: 295-304.
- Schiraldi, C. and M. de Rosa, 2002. The production of biocatalysts and biomolecules from extremophiles. Trends Biotechnol., 20: 515-521.
- van den Burg, B., 2003. Extremophiles as a source for novel enzymes. Curr. Opin. Microbiol., 6: 213-218.
- Eisenmenger, M.J. and J.I. Reyes-De-Corcuera, 2009. High pressure enhancement of enzymes: A review. Enzyme Microbiol. Technol., 45: 331-347.
- Beg, Q.K., V. Sahai and R. Gupta, 2003. Statistical media optimization and alkaline protease production from Bacillus mojavensis in a bioreactor. Process Biochem., 39: 203-209.
- Merheb, C.W., H. Cabral, E. Gomes and R. da-Silva, 2007. Partial characterization of protease from a thermophilic fungus, Thermoascus aurantiacus and its hydrolytic activity on bovine casein. Food Chem., 104: 127-131.
- Coral, G., B. Arikan, M.N. Unaldi and H. Guvenmez, 2003. Thermostable alkaline protease produced by an Aspergillus niger strain. Ann. Microbiol., 53: 491-498.
- Sivasankar, P., S. Sugesh, P. Vijayanand, K. Sivakumar, S. Vijayalakshmi, T. Balasubramanian and P. Mayavu, 2013. Efficient production of L-asparaginase by marine Streptomyces sp. Isolated from bay of Bengal, India. Afr. J. Microbiol. Res., 7: 4015-4021.
- Carrim, A.J.I., E.C. Barbosa and J.D.G. Vieira, 2006. Enzymatic activity of endophytic bacterial isolates of Jacaranda decurrens Cham. (Carobinha-do-campo). Braz. Arch. Biol. Technol., 49: 353-359.
- Miki, Y., H. Ichinose and H. Wariishi, 2010. Molecular characterization of lignin peroxidase from the white-rot basidiomycete Trametes cervina: A novel fungal peroxidase. FEMS Microbiol. Lett., 304: 39-46.
- Borneman, J. and R.J. Hartin, 2000. PCR primers that amplify fungal rRNA genes from environmental samples. Appl. Environ. Microbiol., 66: 4356-4360.
- Ding, F., M. Manosas, M.M. Spiering, S.J. Benkovic, D. Bensimon, J.F. Allemand and V. Croquette, 2012. Single-molecule mechanical identification and sequencing. Nat. Methods, 9: 367-372
- Agrawal, D., P. Patidar, T. Banerjee and S. Patil, 2004. Production of alkaline protease by Penicillium sp. under SSF conditions and its application to soy protein hydrolysis. Process Biochem., 39: 977-981.
- Singh, J., R.M. Vohra and D.K. Sahoo, 2004. Enhancedproduction of alkaline proteases by Bacillus sphaericus using fed-batch culture. Process Biochem., 39: 1093-1101.
- Wang, R.B., J.K. Yang, C. Lin, Y. Zhang and K.Q. Zhang, 2006. Purification and characterization of an extracellular serine protease from the nematode-trapping fungus Dactylella shizishanna. Lett. Appl. Microbiol., 42: 589-594.
- Lucas, A.D. and V.J. Tomazic-Jezic, 2000. Modification of the lowry method for analysis of soluble latex proteins. Toxicol. Method, 10: 165-179.
- Boitano, S., A.N. Flynn, C.L. Sherwood, S.M. Schulz, J. Hoffman, I. Gruzinova and M.O. Daines, 2011. Alternaria alternataserine proteases induce lung inflammation and airway epithelial cell activation via PAR2. Am. J. Physiol. Lung Cell Mol. Physiol., 300: L605-L614.
- Nowakowski, A.B., W.J. Wobig and D.H. Petering, 2014. Native SDS-PAGE: High resolution electrophoretic separation of proteins with retention of native properties including bound metal ions. Metallomics, 6: 1068-1078.
- Sivasubramanian, S., B.M. Manohar, A. Rajaram and R. Puvanakrishnan, 2008. Ecofriendly lime and sulfide free enzymatic dehairing of skins and hides using a bacterial alkaline protease. Chemosphere, 70: 1015-1024.
- Dugan, F.M. and F.M. Dugan, 2006. The Identification of Fungi: An Illustrated Introduction with Keys, Glossary and Guide to Literature. American Phytopathological Society, Saint Paul, MN, USA., ISBN: 9780890543368, Pages: 176.
- Hajji, M., S. Kanoun, M. Nasri and N. Gharsallah, 2007. Purification and characterization of an alkaline serine-protease produced by a new isolated Aspergillus clavatus ES1. Process Biochem., 42: 791-797.
- Wang, S.L., Y.H. Chen, C.L. Wang, Y.H. Yen and M.K. Chern, 2005. Purification and characterization of a serine protease extracellularly produced by Aspergillus fumigatus in a shrimp and crab shell powder medium. Enzyme Microb. Technol., 36: 660-665.
- da Silveira, G.G., G.M. de Oliveira, E.J. Ribeiro, R. Monti and J. Contiero, 2005. Microbial rennet produced by Mucor miehei in solid-state and submerged fermentation. Braz. Arch. Biol. Technol., 48: 931-937.
- Wells, M.L., P. Potin, J.S. Craigie, J.A. Raven and S.S. Merchant et al., 2017. Algae as nutritional and functional food sources: Revisiting our understanding. J. Appl. Phycol., 29: 949-982.
- Salehi, B., J. Sharifi-Rad, A.M.L. Seca, D.C.G.A. Pinto and I. Michalak et al., 2019. Current trends on seaweeds: looking at chemical composition, phytopharmacology, and cosmetic applications. Molecules.
- Khotimchenko, S.V., V.E. Vaskovsky and T.V. Titlyanavo, 2002. Fatty acids of marine algae from the pacific coast of North California. Bot. Mar., 45: 17-22.
- Tremacoldi, C.R. and E.C. Carmona, 2005. Production of extracellular alkaline proteases by Aspergillus clavatus. World J. Microbiol. Biotechnol., 21: 169-172.
- Sharma, K.M., R. Kumar, S. Panwar and A. Kumar, 2017. Microbial alkaline proteases: Optimization of production parameters and their properties. J. Genet. Eng. Biotechnol., 15: 115-126.
- Shanthi, G., M. Premalatha and N. Anantharaman, 2021. Potential utilization of fish waste for the sustainable production of microalgae rich in renewable protein and phycocyanin-Arthrospira platensis/Spirulina. J. Cleaner Prod.
- Espósito, T.S., I.P.G. Amaral, D.S. Buarque, G.B. Oliveira, L.B. Carvalho Jr. and R.S. Bezerra, 2009. Fish processing waste as a source of alkaline proteases for laundry detergent. Food Chem., 112: 125-130.
- Shih, I.L., L.G. Chen, T.S. Yu, W.T. Chang and S.L. Wang, 2003. Microbial reclamation of fish processing wastes for the production of fish sauce. Enzyme Microb. Technol., 33: 154-162.
- Bhaskar, N. and N.S. Mahendrakar, 2008. Protein hydrolysate from visceral waste proteins of Catla (Catla catla): Optimization of hydrolysis conditions for a commercial neutral protease. Bioresour. Technol., 99: 4105-4111.
- Bhaskar, N., T. Benila, C. Radha and R.G. Lalitha, 2008. Optimization of enzymatic hydrolysis of viscera proteins of Catla (Catla catla) for preparing protein hydrolysate using a commercial protease. Bioresour. Techol., 99: 335-343.
- Habicher, T., E.K.A. Rauls, F. Egidi, T. Keil, T. Klein, A. Daub and J. Büchs, 2020. Establishing a fed-batch process for protease expression with Bacillus licheniformis in polymer-based controlled‐release microtiter plates. Biotechnol. J.
- Olivieri, F., M.E. Zanetti, C.R. Oliva, A.A. Covarrubias and C.A. Casalongué, 2002. Characterization of an extracellular serine protease of Fusarium eumartii and its action on pathogenesis related proteins. Eur. J. Plant Pathol., 108: 63-72.
- Kim, J.D., 2004. Purification and characterization of an extracellular alkaline protease from Aspergillus niger C-15. Mycobiology, 32: 74-78.
- Charles, P., V. Devanathan, P. Anbu, M.N. Ponnuswamy, P.T. Kalaichelvan and B.K. Hur, 2008. Purification, characterization and crystallization of an extracellular alkaline protease from Aspergillus nidulans HA-10. J. Basic Microbiol., 48: 347-352.
- Tunga, R., B. Shrivastava and R. Banerjee, 2003. Purification and characterization of a protease from solid state cultures of Aspergillus parasiticus. Process Biochem., 38: 1553-1558.
- Tremacoldi, C.R., R. Monti, H.S. Selistre-De-Araújo and E.C. Carmona, 2007. Purification and properties of an alkaline protease of Aspergillus clavatus. World J. Microbiol. Biotechnol., 23: 295-299.
- Chi, Z. and S. Zhao, 2003. Optimization of medium and cultivation conditions for pullulan production by a new pullulan-producing yeast strain. Enzyme Microb. Technol., 33: 206-211.
- Uyar, F. and Z. Baysal, 2004. Production and optimization of process parameters for alkaline protease production by a newly isolated Bacillus sp. under solid state fermentation. Process Biochem., 39: 1893-1898.
- Venugopal, M. and A.V. Saramma, 2006. Characterization of alkaline protease from Vibrio fluvialis strain VM10 isolated from a mangrove sediment sample and its application as a laundry detergent additive. Process Biochem., 41: 1239-1243.
- Han-Seung, J., G.C. Park, K.T. Kim, S.R. Paik and C.S. Chang, 2001. Simple methods for alkaline protease purification from the polychaeta, Periserrula leucophryna. Process Biochem., 37: 299-303.
- Banik, R.M. and M. Prakash, 2004. Laundry detergent compatibility of the alkaline protease from Bacillus cereus. Microbiol. Res., 159: 135-140.
- Rao, C.S., T. Sathish, P. Ravichandra and R.S. Prakasham, 2009. Characterization of thermo- and detergent stable serine protease from isolated Bacillus circulans and evaluation of eco-friendly applications. Process Biochem., 44: 262-268.
- Savitha, S., S. Sadhasivam, K. Swaminathan and F.H. Lin, 2011. Fungal protease: Production, purification and compatibility with laundry detergents and their wash performance. J. Taiwan Inst. Chem. Eng., 42: 298-304.
- Sundararajan, S., C.N. Kannan and S. Chittibabu, 2011. Alkaline protease from Bacillus cereus VITSN04: Potential application as a dehairing agent. J. Biosci. Bioeng., 111: 128-133.
- Shrinivas, D. and G.R. Naik, 2011. Characterization of alkaline thermostable keratinolytic protease from thermoalkalophilic Bacillus halodurans JB 99 exhibiting dehairing activity. Int. Biodeterior. Biodegrad., 65: 29-35.
How to Cite this paper?
APA-7 Style
Sugesh,
S., Palaniyappan,
S., Marimuthu,
N., Mani,
S., Packiyam,
M. (2023). Purification and Characterization of an Alkaline Protease from Aspergillus fumigatus Cas1 Isolated from Industrial Polluted Area. Asian Journal of Biological Sciences, 16(1), 60-70. https://doi.org/10.3923/ajbs.2023.60.70
ACS Style
Sugesh,
S.; Palaniyappan,
S.; Marimuthu,
N.; Mani,
S.; Packiyam,
M. Purification and Characterization of an Alkaline Protease from Aspergillus fumigatus Cas1 Isolated from Industrial Polluted Area. Asian J. Biol. Sci 2023, 16, 60-70. https://doi.org/10.3923/ajbs.2023.60.70
AMA Style
Sugesh
S, Palaniyappan
S, Marimuthu
N, Mani
S, Packiyam
M. Purification and Characterization of an Alkaline Protease from Aspergillus fumigatus Cas1 Isolated from Industrial Polluted Area. Asian Journal of Biological Sciences. 2023; 16(1): 60-70. https://doi.org/10.3923/ajbs.2023.60.70
Chicago/Turabian Style
Sugesh, Shanmugam, Sivasankar Palaniyappan, Nandhini Marimuthu, Suriya Mani, and Mayavu Packiyam.
2023. "Purification and Characterization of an Alkaline Protease from Aspergillus fumigatus Cas1 Isolated from Industrial Polluted Area" Asian Journal of Biological Sciences 16, no. 1: 60-70. https://doi.org/10.3923/ajbs.2023.60.70

This work is licensed under a Creative Commons Attribution 4.0 International License.



