Evaluation of CDK6 Inhibitory Potential of 16-(Substituted)-Dehydroepiandrosterone Derivatives Using Computational Tools
| Received 13 Apr, 2023 |
Accepted 15 Jul, 2023 |
Published 30 Sep, 2023 |
Background and Objective: The use of corticosteroids in cancer treatment is common, they are especially effective in individuals with substantial peritumoral edema and neurological impairments. The present study aimed to correlate the 57 novel steroidal derivatives with their ability to inhibit CDK6 protein which is an important target for brain cancer. Materials and Methods: The steroidal molecules and their activities were taken from the literature. The molecules were drawn and energies were minimised. The molecular docking studies were performed using the protein CDK6(PDB-6OQO). The 16-substituted epiandrosterone derivatives were evaluated for their binding potential to the protein using AutoDock. Results: The best-docked compound (S228) is showing hydrogen bonding with LYSA: 43, LYSA29 and ASPA: 163, one pi-sigma bond with ALAA: 162 and alkyl binding with LYSA: 29, VALA: 27, ILEA: 19, ILUA: 152. The derivatives possessing 16-(4-nitrobenzylidene) substitution have exhibited higher binding energies in comparison to other substituents. Conclusion: These dehydroepiandrosterone derivatives have exhibited good binding energies for CDK6 protein. The studies have revealed that at position 3, the substitution of electro-negative will enhance the affinity of the molecules towards the protein.
| Copyright © 2023 Dubey et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Corticosteroids are commonly used in cancer treatment and they are especially effective in individuals with substantial peritumoral edema and neurological impairments. Despite their extensive history of usage and enormous influence in clinical oncology over several decades, little is understood about the molecular and clinical processes by which corticosteroids work1,2.
Steroids were first used in the treatment of brain cancer patients approximately 50 years ago because of their potent anti-edema properties. In 1952, ingraham was the first to utilize cortisone to treat postoperative cerebral edoema in neurosurgical patients3 and in 1957, kofman was the first to use prednisone for peritumoral edoema caused by brain metastases4. Dexamethasone was shown to efficiently relieve cerebral edoema caused by brain tumors more than 40 years ago, which changed brain tumor patient care. Galicich’s research found that large doses of dexamethasone can prevent the growth of brain tumours1.
Dexamethasone has continued to be the most helpful treatment for those with brain cancer. When compared to other corticosteroids, it has a reduced sodium and water retention index, which has a clear benefit for the treatment5. Methylprednisolone, dexamethasone, prednisone and prednisolone have since been shown to have anticancer properties against hematologic cancers6. In cancer patients, glucocorticoids have also been shown to help decrease tumor-related discomfort, reduce nausea and vomiting and improve appetite7. Importantly, in individuals with carcinomatous meningitis and CNS lymphoma, steroids are frequently prescribed8. Because of its extended half-life, low mineralocorticoid action and minimal likelihood to generate psychosis, dexamethasone has become the medication of choice in neuro-oncology2.
Despite its widespread use, only a few prospective clinical trials to assess the appropriate dose of dexamethasone in brain cancer patients have been conducted9,10. The effectiveness of steroids in decreasing tumor-related edema has been thoroughly established11,12.
Glioblastoma is one of the most malignant types of brain tumour. Currently, not much of the treatment is available for it. Patients are usually treated with radiation along with temozolomide (TMZ) therapy. But the effect is generally short duration and there is a high degree of reoccurrence. Dehydroepiandrosterone (DHEA), is a neurosteroid that maintains the health of neurons and astrocytes. DHEA stops tmz-induced apoptosis by reducing DNA damage in MGMT (O6-methylguanine-DNA methyl-transferase)-deficient glioblastoma. The exact mechanism for the same is yet to be established13-15.
An essential enzyme, cyclin-dependent kinase 6 (CDK6) controls the cell cycle progression in the body. Recently, it has also been demonstrated that CDK6 plays a transcriptional part in tumour angiogenesis. In particular, up-regulated CDK6 activity is linked to the development of many cancer types, especially CNS cancer16.
In the present study, the CDK6 inhibitory potential of some novel steroidal molecules. In silico binding energies of 57 novel 16-substituted DHEA derivatives as CDK6 inhibitors were calculated and protein interactions were estimated.
MATERIALS AND METHODS
Study area: The present study was conducted at College of Pharmaceutical Sciences, Dayananda Sagar University, Bengaluru. The study was carried out from April, 2022 to August, 2022.
Collection of the data: The data was collected from the synthesis and biological evaluation of a series of 16-substituted DHEA derivatives from literature17-19. These compounds were found to have potent anti-cancer activity, particularly against cancer cells of the Central Nervous System (CNS). The 57 novel 16-substituted DHEA derivatives were first drawn in 2D using chemical drawing software. The structures were then cleaned up to remove any errors or inconsistencies. Next, the force field MM2 was used to perform energy minimization on the structures. This process helped to ensure that the structures were in their most stable form. Finally, the structures were saved in PDB format, which is a standard file format for storing protein and ligand structures.
The CDK6 protein ID 6OQO was selected from the Protein Data Bank (PDB). The CDK6 protein that has been selected has a resolution of 1.98 Å, which is considered to be high resolution. The protein is also a non-mutant variant from homo sapiens, which means that it is the same protein that is found in humans. This was considered to be appropriate for the current studies because it’s important to investigate the potential of the 16-substituted DHEA derivatives to inhibit CDK6 in humans. The 16-substituted DHEA derivatives were prepared in pdb format, which is the same file format as the CDK6 protein. The water
molecules were removed from the protein structure to create a pdbqt file, which is a format that is specifically designed for molecular docking studies. The x, y and z coordinates of the CDK6 protein were calculated to determine the size of the grid that would be used for the molecular docking studies. The co-crystallized molecule was removed from the protein structure to ensure that the docking results were not biased by the presence of the other molecule. Finally, AutoDock 4.0 was used to perform the molecular docking studies. AutoDock 4.0 is a software program that is used to predict the binding affinity of ligands to proteins.
RESULTS AND DISCUSSION
The molecular docking studies of the novel 16-substituted DHEA derivatives were performed to see the binding abilities of these molecules with CDK6 protein 6OQO, to act as inhibitors. Table 1 shows the comprehensive results of the structures of steroidal derivatives, their h-bonding interaction with specific amino acids in the protein along with the binding energies of the top 27 compounds. The results were compared with the standard drug dexamethasone.
| Table 1: | Results of molecular docking studies of 16-substituted dehydroepiandrosterone derivatives with CDK6 (PDB-6OQO) macromolecule | |||
| Code | Binding energy | Bonding | Interacting amino acids |
| S228 | -11.69 | 2 H-bond | LYSA: 43, ASPA: 163, LYSA: 29 |
| pi-Sigma | ALAA: 162 | ||
| pi-Alkyl | LYSA: 29, VALA: 27, ILEA: 19, ILUA: 152 | ||
| S227 | -11.08 | 1H-bond | TYRA: 24 |
| pi-Sigma | ALAA: 162 | ||
| pi-Alkyl | ILEA: 19, LEUA: 152, VALA: 27 | ||
| S224 | -11.07 | 2H-bond | ASPA: 102, LYSA: 43 |
| pi-Sigma | VALA: 27 | ||
| pi-Alkyl | ALAA: 162, ILEA: 19, LEUA: 152 | ||
| S268 | -10.89 | 2 H-bond | LYSA: 43, ASPA: 104 |
| pi-Sigma | ALAA: 162 | ||
| pi-Alkyl | VALA: 77, ALAA: 41, VALA: 27, ILEA: 19, VALA: 101, LEUA: 152 | ||
| S256 | -10.71 | 4H-bond | LYSA: 43, GLNA: 149, THRA: 182, VALA: 181 |
| pi-Sigma | ALAA: 162 | ||
| pi-Alkyl | LEUA: 152, VALA: 27, TYRA: 24 | ||
| S241 | -10.22 | 2H-bond | TYRA: 24, ASPA: 102 |
| pi-Sigma | PHEA: 98, VALA: 27 | ||
| pi-Alkyl | ALAA: 162, ILEA: 19 | ||
| S250 | -10.2 | 2H-bond | TYRA: 24, ASPA: 102 |
| pi-Sigma | ALAA: 162, PHEA: 98 | ||
| pi-Alkyl | VALA: 27, ALAA: 41, VALA: 77, ILEA: 19, LEUA: 152 | ||
| S205 | -9.69 | 1H-bond | ASPA: 102 |
| pi-Sigma | ALAA: 162, PHEA: 98 | ||
| pi-Alkyl | VALA: 27, ALAA: 41, VALA: 77, ILEA: 19, LEUA: 152 | ||
| S265 | -9.75 | 3H-bond | LYSA: 43, HISA: 100, LYSA: 29 |
| pi-Alkyl | LEUA: 152, ALAA: 162, ILEA: 19, LEUA: 152, VALA: 27 | ||
| S236 | -9.44 | 1H-bond | HISA: 100 |
| pi-Sigma | VALA: 27 | ||
| pi-Alkyl | ALAA: 41, PHEA: 98, ILEA: 19, LEUA: 152 | ||
| S267 | -9.36 | 1H-bond | LYSA: 111 |
| pi-Sigma | LEUA: 152 | ||
| pi-Alkyl | ILEA: 19, LYSA: 29, HISA: 100, VALA: 27, PHEA: 98, ALAA: 162, | ||
| VALA: 101, ALAA: 41 | |||
| S261 | -9.27 | 2H-bond | LYSA: 43, TYRA: 24 |
| pi-Sigma | VALA: 27 | ||
| pi-Alkyl | ILEA: 19, ALAA: 41, VALA: 77, LEUA: 152, PHEA: 98, TYRA: 24, | ||
| AlAA: 162 | |||
| S245 | -9.14 | 1H-bond | LYSA: 43 |
| pi-Sigma | VALA: 27 | ||
| pi-Alkyl | ALAA: 162, LEUA: 152 | ||
| S251 | -9.03 | 1H-bond | VALA: 27 |
| py-Alkyl | ILEA: 19, ASPA: 104 | ||
| S223 | -8.92 | 1H-bond | LYSA: 43 |
| pi-Sigma | TYRA: 24, PHEA: 98, ALAA: 162 | ||
| pi-Alkyl | LEUA: 152, VALA: 27, ILEA: 19 | ||
| S262 | -8.89 | 2H-bond | TYRA: 24, ASPA: 102 |
| pi-Sigma | ALAA-162 | ||
| pi-Alkyl | TYRA: 24, ILEA: 19, LEUA: 152, HISA: 100, VALA: 77 | ||
| S266 | -8.87 | 1H-bond | HISA: 100 |
| pi-Alkyl | ALAA: 162, ILEA: 19, VALA: 27 | ||
| S264 | -8.86 | 3H-bond | TYRA: 24, LYSA: 43, ASPA: 102 |
| pi-Alkyl | ILEA: 19, VALA: 27, ALAA: 162, ILEA: 19 | ||
| S196 | -8.54 | pi-Sigma | TYRA: 24 |
| pi-Alkyl | ALAA: 162, ALAA: 41, ILEA: 19, VLA: 27, VLA: 101, VLA: 77, | ||
| ALAA: 162, LEUA: 152, PHEA: 98, GLUA: 99, ASPA: 104, | |||
| GLNA: 149, ASNA: 150 | |||
| S243 | -8.46 | pi-Sigma | LEUA: 152 |
| pi-Alkyl | ALAA: 41, ILEA: 19, VALA: 27, ALAA: 162 | ||
| S207 | -8.35 | 1H-bond | LYSA: 29 |
| pi-Sigma | TYRA: 24, ILEA: 19 | ||
| pi-Alkyl | IEUA: 152, VALA: 27 | ||
| S247 | -8.32 | 2H-bond | LYSA: 29, VALA: 101 |
| pi-Alkyl | TYRA: 24, VALA: 27, LEUA: 152 | ||
| S197 | -8.16 | 1H-bond | HISA: 100 |
| py-Sigma | ILEA: 19, LEUA: 152 | ||
| py-Alkyl | ALAA: 41, VALA: 27 | ||
| S230 | -7.93 | 2H-bond | ALAA: 41, ILEA: 19 |
| py-Alkyl | VALA: 27 | ||
| S263 | -7.92 | 1H-bond | LYSA: 43 |
| py-Alkyl | TYRA: 24, VALA: 27, LEUA: 152 | ||
| S188 | -7.86 | py-Alkyl | VALA: 27, VALA: 77, ILEA: 19, LEUA: 152, ALA: 162, ALAA: 41, |
| TYRA: 24, PHEA: 98 | |||
| S184 | -7.11 | py-Alkyl | VAL: 1, 01 VAL: 27, ALA: 41, ILEA: 19, HIS: 100 |
| 0H-bond | |||
| DEXAMETHASONE | -7.45 | 3H-bond | ILEA: 19, ILEA: 19, LYSA: 43 |
| py-Alkyl | ALAA: 162, LEUA: 152, TYRA: 24 |
It is evident from the results that the steroid derivatives possessing 16-(4-nitrobenzylidene) substituted (compound No. S-228, 227, 224, 268 and 256) are exhibiting higher binding energies in comparison to other substituents. At position 3, the substitution of acetate group, amines or pyrrole ring is preferred.
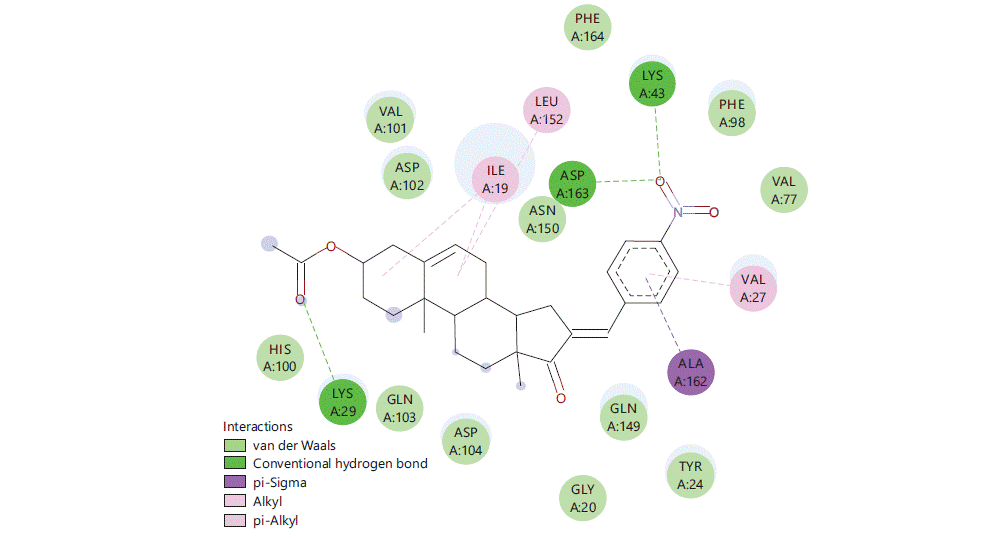
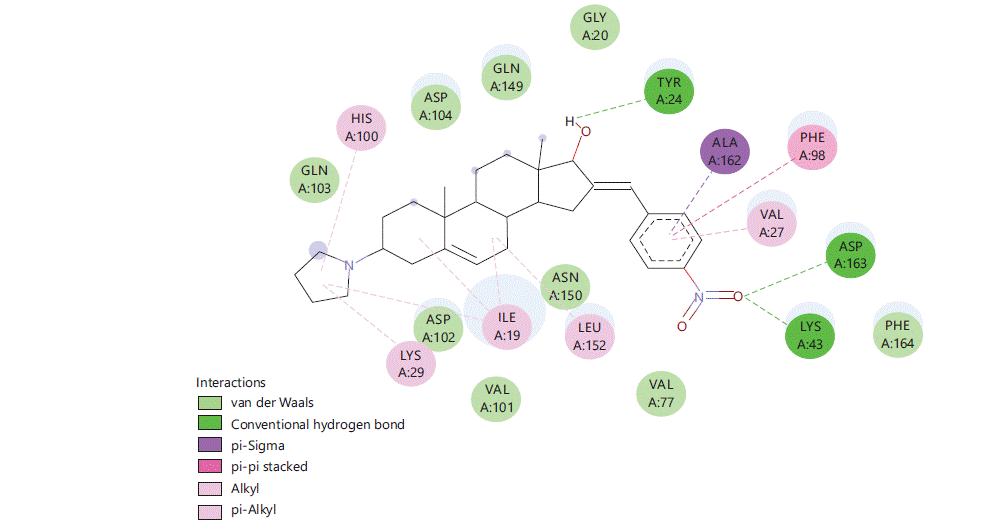
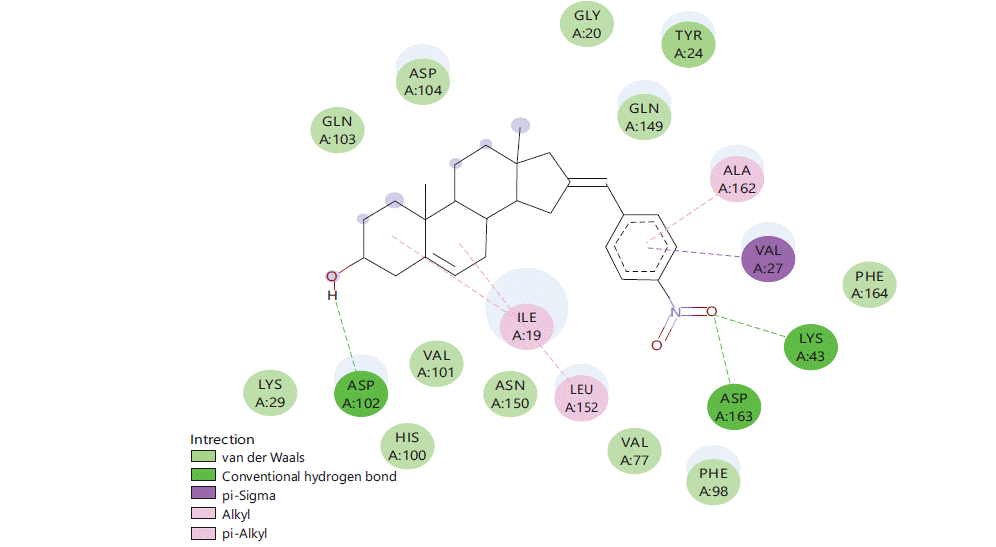
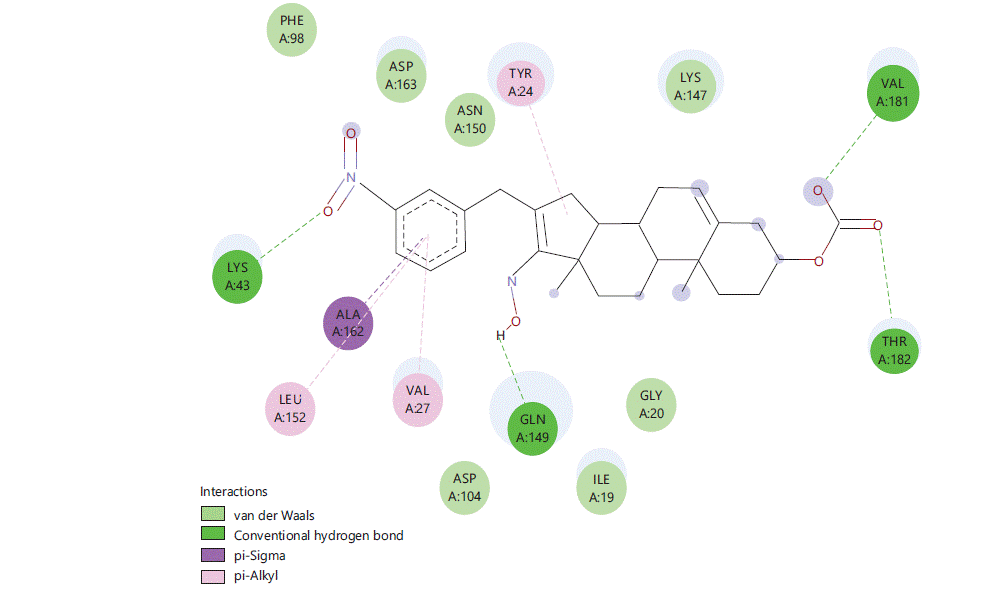
Figure 1 to 5 shows the binding interactions of the five best steroidal derivatives.
The best-docked compound (S228) is showing 2H-bond bonding (LYSA: 43, LYSA29 and ASPA: 163), one pi-sigma bond (ALAA: 162) and alkyl binding with (LYSA: 29, VALA: 27, ILEA: 19 and ILUA: 152) some of the amino acids are even showing van der waals interactions.
Similar kind of interactions are visible in other steroidal derivatives exhibiting good binding interactions (Fig. 2-5).
Almost all steroidal derivatives exhibit better binding energy and interactions compared to the standard drug-dexamethasone. There are fewer studies that have been done on the use of steroids as an anticancer agent20-22 an in silico study conducted by Saikia et al.23 on marine steroids has shown their potential to inhibit CDK4 and Bcl-2 receptors. The investigation of steroidal molecules against CDK6 inhibition has not been explored previously. In this aspect based on the results obtained from this study, it can be said that the steroidal molecules do possess the potential to bind with CDK6 receptor through amino acids LYSA: 43, TYRA: 24, VALA: 27 and LEUA: 152. These amino acids can be potential binding sites for these steroidal molecules to exhibit anticancer activity in the CNS. When compared 16 (4-nitro benzylidene) substitution has proven good for the binding of the molecule in the active site of CDK6 protein (PDB-6OQO). Hence, the introduction of electronegative groups like nitro, acetate and amines may prove beneficial for the designing of potent steroidal derivatives possessing activity for CNS cancer.
|
|
|

|
|
CONCLUSION
In conclusion, this study investigated a new target for steroidal molecules and the relationship between steroidal derivatives and their potential to inhibit CDK6 receptors in silico has been established. Various 16-substituted DHEA derivatives were evaluated. The results of the study have shown that the introduction of an electronegative group at the 16-position of steroidal molecules can enhance their binding to CDK6 receptors through amino acids LYSA: 43, TYRA: 24, VALA: 27 and LEUA: 152. This finding has important implications for the development of new drugs that target CDK6. Further work in this direction is warranted to optimize the binding affinity and selectivity of these compounds.
SIGNIFICANCE STATEMENT
This study has been conducted to evaluate the potential of lesser explored-substituted dehydroepiandrosterone derivatives as CDK6 inhibitors for their potential use as CNS anticancer molecules. These studies have revealed that these steroidal structures possess good binding affinity towards the 6OQO protein. Further detailed studies in this direction can give a lead compound.
REFERENCES
- Jessurun, C.A.C., A.F.C. Hulsbergen, L.D. Cho, L.S. Aglio, R.D.S.N. Tewarie and M.L.D. Broekman, 2019. Evidence-based dexamethasone dosing in malignant brain tumors: What do we really know? J. Neuro-Oncol., 144: 249-264.
- Ryken, T.C., M. McDermott, P.D. Robinson, M. Ammirati and D.W. Andrews et al., 2010. The role of steroids in the management of brain metastases: A systematic review and evidence-based clinical practice guideline. J. Neuro-Oncol., 96: 103-114.
- Ingraham, F.D., D.D. Matson and R.L. McLaurin, 1952. Cortisone and ACTH as an adjunct to the surgery of craniopharyngiomas. New Engl. J. Med., 246: 568-571.
- Kofman, S., J.S. Garvin, D. Nagamani and S.G. Taylor, 1957. Treatment of cerebral metastases from breast carcinoma with prednisolone. JAMA, 163: 1473-1476.
- Bell, B.A., D.M. Kean, H.L. Macdonald, G.H. Barnett and R.H.B. Douglas et al., 1987. Brain water measured by magnetic resonance imaging: Correlation with direct estimation and changes after mannitol and dexamethasone. Lancet, 329: 66-69.
- Inaba, H. and C.H. Pui, 2010. Glucocorticoid use in acute lymphoblastic leukaemia. Lancet Oncol., 11: 1096-1106.
- Markman, M., V. Sheidler, D.S. Ettinger, S.A. Quaskey and E.D. Mellits, 1984. Antiemetic efficacy of dexamethasone-randomized, double-blind, crossover study with prochlorperazine in patients receiving cancer chemotherapy. New Engl. J. Med., 311: 549-552.
- Todd, F.D., C.A. Miller, A.J. Yates and L.J. Mervis, 1986. Steroid-induced remission in primary malignant lymphoma of the central nervous system. Surg. Neurol., 26: 79-84.
- Vecht, C.J., A. Hovestadt, H.B.C. Verbiest, J.J. van Vliet and W.L.J. van Putten, 1994. Dose‐effect relationship of dexamethasone on Karnofsky performance in metastatic brain tumors: A randomized study of doses of 4, 8, and 16 mg per day. Neurology, 44: 675-680.
- Graham, P.H., A. Capp, G. Delaney, G. Goozee and B. Hickey et al., 2006. A pilot randomised comparison of dexamethasone 96mg vs 16mg per day for malignant spinal-cord compression treated by radiotherapy: TROG 01.05 superdex study. Clin. Oncol., 18: 70-76.
- Marantidou, A., C. Levy, A. Duquesne, R. Ursu and O. Bailon et al., 2010. Steroid requirements during radiotherapy for malignant gliomas. J. Neuro-Oncol., 100: 89-94.
- Miller, J.D. and P. Leech, 1975. Effects of mannitol and steroid therapy on intracranial volume-pressure relationships in patients. J. Neurosurg., 42: 274-281.
- Miller, J.D., R. Sakalas, J.D. Ward, H.F. Young, W.E. Adams, J.K. Vries and D.P. Becker, 1977. Methylprednisolone treatment in patients with brain tumors. Neurosurgery, 1: 114-117.
- Yang, W.B., J.Y. Chuang, C.Y. Ko, W.C. Chang and T.I. Hsu, 2019. Dehydroepiandrosterone induces temozolomide resistance through modulating phosphorylation and acetylation of Sp1 in glioblastoma. Mol. Neurobiol., 56: 2301-2313.
- Tadesse, S., M. Yu, M. Kumarasiri, B.T. Le and S. Wang, 2015. Targeting CDK6 in cancer: State of the art and new insights. Cell Cycle, 14: 3220-3230.
- Yeung, W.T.I., T.Y. Lee, R.F. Del Maestro, R. Kozak, J. Bennett and T. Brown, 1994. Effect of steroids on iopamidol blood-brain transfer constant and plasma volume in brain tumors measured with X-ray computed tomography. J. Neuro-Oncol., 18: 53-60.
- Dubey, S., D.P. Jindal and P. Piplani, 2005. Synthesis and antineoplastic activity of some 16-benzylidene substituted steroidal oximes. Indian J. Chem., 44: 2126-2137.
- Dubey, S., P. Piplani and D.P. Jindal, 2004. Synthesis and in vitro antineoplastic evaluation of certain 16-(4-substituted benzylidene) derivatives of androst-5-ene. Chem. Biodivers., 1: 1529-1536.
- Dubey, S., P. Piplani and D. Jindal, 2005. Synthesis and evaluation of some 16-benzylidene substituted 3,17-dioximino androstene derivatives as anticancer agents. Lett. Drug Des. Discovery, 2: 537-545.
- Elmegeed, G.A., S.M.M. Yahya, M.M. Abd-Elhalim, M.S. Mohamed, R.M. Mohareb and G.H. Elsayed, 2016. Evaluation of heterocyclic steroids and curcumin derivatives as anti-breast cancer agents: Studying the effect on apoptosis in MCF-7 breast cancer cells. Steroids, 115: 80-89.
- Tolmacheva, I.A., A.V. Nazarov, D.V. Eroshenko and V.V. Grishko, 2018. Synthesis, cytotoxic evaluation, and molecular docking studies of the semi-synthetic “triterpenoid-steroid” hybrids. Steroids, 140: 131-143.
- Bhattacharya, A., P.S. Guha, N. Chowdhury, A. Bagchi and D. Guha, 2023. Virtual screening and molecular docking of flavone derivatives as a potential anticancer drug in the presence of dexamethasone. Biointerface Res. Appl. Chem., 13.
- Saikia, S., B. Kolita, P.P. Dutta, D.J. Dutta and Neipihoi et al., 2015. Marine steroids as potential anticancer drug candidates: In silico investigation in search of inhibitors of Bcl-2 and CDK-4/Cyclin D1. Steroids, 102: 7-16.
How to Cite this paper?
APA-7 Style
Dubey,
S., Goni,
T., Maheshwari,
T., Kumar,
V. (2023). Evaluation of CDK6 Inhibitory Potential of 16-(Substituted)-Dehydroepiandrosterone Derivatives Using Computational Tools. Asian Journal of Biological Sciences, 16(3), 401-408. https://doi.org/10.3923/ajbs.2023.401.408
ACS Style
Dubey,
S.; Goni,
T.; Maheshwari,
T.; Kumar,
V. Evaluation of CDK6 Inhibitory Potential of 16-(Substituted)-Dehydroepiandrosterone Derivatives Using Computational Tools. Asian J. Biol. Sci 2023, 16, 401-408. https://doi.org/10.3923/ajbs.2023.401.408
AMA Style
Dubey
S, Goni
T, Maheshwari
T, Kumar
V. Evaluation of CDK6 Inhibitory Potential of 16-(Substituted)-Dehydroepiandrosterone Derivatives Using Computational Tools. Asian Journal of Biological Sciences. 2023; 16(3): 401-408. https://doi.org/10.3923/ajbs.2023.401.408
Chicago/Turabian Style
Dubey, Sonal, Tilak Goni, Tanya Maheshwari, and Vedaanshu Kumar.
2023. "Evaluation of CDK6 Inhibitory Potential of 16-(Substituted)-Dehydroepiandrosterone Derivatives Using Computational Tools" Asian Journal of Biological Sciences 16, no. 3: 401-408. https://doi.org/10.3923/ajbs.2023.401.408

This work is licensed under a Creative Commons Attribution 4.0 International License.



