Evaluation of Genotype Coefficient of Variation (GCV) and Phenotypic Coefficient of Variation (PCV) of Seed Yield in Sesamum indicum Cultivars
| Received 01 May, 2024 |
Accepted 04 Aug, 2024 |
Published 31 Dec, 2024 |
Background and Objective: Performance is a complex trait influenced by various factors that are inherited in a polygenic manner and highly subject to environmental changes. Therefore, functional components that are less affected by the environment should be selected. The aim of this study was to evaluate the genetic parameters of seed yield in sesame cultivars. Materials and Methods: In addition, Genotype Coefficient of Variation (GCV) and Phenotypic Coefficient of Variation (PCV), heritability, genetic progress, the study of correlation between performance and its components and their relative contribution to performance in breeding programs are of great importance. Results: The results showed that the heritability of all traits was high, except for day to maturity, which had moderate heritability. The General Average (GA) was high for all traits except weight of 1,000 seeds, days to 50% flowering, capsule length, protein content, oil content and days to maturity. Conclusion: The study concludes that southern region of Ethiopia showed the highest seed yield. The heritability of all traits was high, except for day to maturity, which had moderate heritability.
| Copyright © 2024 Ibrahim and Tahmasbe. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Sesame plant with the scientific name (Sesamum Indicum L) is from the pedaliaceae family and is one of the oldest valuable agricultural plants. Sesame is one of our prehistoric oil plants that dates back to 3050-3500 BC (Before the Christ) and was cultivated in ancient times. Sesame is widely cultivated in the world and the main areas of sesame production are tropical and subtropical regions of the world. About 20 species of wild sesame cultivated in Asia and Africa, the Indian species has a high economic value1,2. The superior quality of sesame oil, which is attributed to the saturated fatty acids as well as unique natural antioxidant activity, distinguishes it from other oilseed products3,4.
Sesame is a thermophile plant that is compatible with the arid- and semi-arid regions of the world and needs sunny weather during the growth season5,6. Given the compatibility of sesame plant with different climates and tolerance of this plant to unpleasant environmental conditions, various experiments are carried out for the expansion of its cultivation7. Therefore, production and development of new varieties of sesame always continue. It has always been at the focal attention of breeders of crops in the world8,9. Existence of genetic diversity in members of the population is one of the criteria for genetic improvement10,11. Determining the breeding methods and optimal selection significantly increases the chance of genetic improvement. The weight of 1,000 seeds, capsules/plant, number of branches/plant and plant height were selected to improve seed yield/plant. The high heritability and high genetic progress for these traits showed that the function of high-value additive genes and phenotypic selection can be effective in improving these traits12,13.
Based on previous studies, it has been determined that the investigated sesame genotypes have significant genetic diversity, which allows effective selection in these genotypes. Also, some of the traits they studied included days up to 50% flowering, plant height, length of fruiting area and number of branches per plant were effective in predicting seed weight per plant because they had the highest heritability with genetic improvement as a percentage14. Considering the importance of sesame due to its high nutritional value for humans, the need for research such as the current proposal to improve the performance of sesame is strongly felt. Thus, the aim of this study was to evaluate of genetic parameters of seed yield in sesame cultivars.
MATERIALS AND METHODS
The present research was applied on (14/5/2022) and (22/3/2023) at Agricultural Research Station of the School of Agriculture, Wasit University, Iraq. The experimental farm has coordinates of a Longitude of 45° and 50 min 33.5 sec East and Latitude of 32° and 29 min 49.8 sec North. To determine the physical and chemical characteristics of the farm soil before cultivation, sampling will be done from 6 different points of the land and randomly from a depth of 0 to 30 cm. Also, the meteorological characteristics of the station, including temperature, relative humidity and rainfall, were measured during the months of the experiment. The experiment was conducted in the form of randomized complete blocks with three replications. Experimental treatments include 16 sesame cultivars (12 Iranian genentypes have been used including Naz tak shakheh, Dashtestan, Dashtestan2, Mahali Mehran, Nashekhifa, Holeil, Shevin, Darab1, TC-25, Yellow white, Darab14, Oltan, Darab2 and 2 genentypes cultivars Iraqi White and Red Iraqi).
The steps of land preparation were done in summer (from June to September), including plowing, land leveling and ditching. Each experimental unit is as long as 4 m and treatments were cultivated in each experimental unit consisting of 4 cultivation lines with a distance of 60 cm from each other and 20 cm plant spacing, which included 48 plots in total. The seeds were planted on the lines. During the growth of the plant, nitrogen fertilizer was given in three stages in the form of a stream along with the first irrigation, stemming and flowering. Necessary care including weed and pest control was also done during the plant growth period.
The investigation traits including number of branches per plant, capsule length, weight of 1000 seeds (g), seed weight per plant (g) and seed yield (kg per hectare) will be measured.
Variance components including additive variance (D), mean and covariance of traits and additive and non-additive effects of genes should be investigated.
Statistical analysis: The statistical analysis of the data was done using Minitab (version 18) and SAS (version 9.1) statistical software and comparing the average data using the LSD test at the 5% probability level. Excel software is used to draw graphs.
Number of buds per plant: The number of buds of cultivars was counted based on observation and randomly from three replicates, five plants from each replicate.
Amount of chlorophyll: The amount of chlorophyll in the leaves of each plant was extracted using the usual methods and measured by a chlorophyll meter (SPAD index).
Amount of chlorophyll was measured using ARNON method: Half a gram of fresh vegetable matter was poured into a Chinese mortar, then grind it using liquid nitrogen and crushed well. The 20 mL of 80% acetone was added to the sample and then put in a centrifuge device at a speed of 6000 rpm (round per minute) for 10 min. The separated upper extract was transferred from the centrifuge to a glass flask.
Some of the samples were poured inside the balloon into the cuvette of a spectrophotometer and then read the absorbance value separately at the wavelengths of 663 nm for chlorophyll a, 645 nm for chlorophyll b and 470 for carotenoids by spectrophotometer. Finally, the amount of chlorophyll a, b and carotenoids was obtained in terms of milligrams per gram of fresh weight of the sample using the following formulas6:
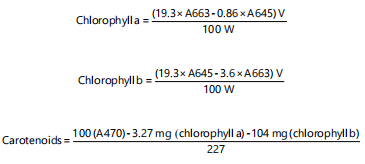 |
| V | = | Volume of strained solution (upper solution obtained from centrifugation) | |
| A | = | Absorption of light at wavelengths of 663, 645 and 470 nm | |
| W | = | Fresh weight of the sample in grams |
Number of branches in the bush: The number of branches of each selected plant was randomly counted and written in the data table. The plant height of the cultivars was evaluated based on centimeters and with the help of meters and randomly from three replicates with 5 plants from each replicate.
Leaf surface was measured using a CI-203 handheld device: A highly portable laser scanner is suitable for fast and non-destructive measurement of leaf surface at any location. Measurements are easily done by relocating the scanner over the leaf. The plant height of the cultivars was evaluated based on centimeters and with the help of meters and randomly from three replicates with five plants from each replicate.
Performance of each plant: The performance of each plant was evaluated randomly from three repetitions, with five plants per repetition. The seeds of the sample were weighed after separation and entered in the data table.
Performance of whole plant per hectare: The performance of each plant was randomly evaluated from 3 repetitions, with 5 plants per repetition. The seeds of the sample were weighed after separation and the number of plants per square meter was converted into hectares and entered in the data table.
RESULTS AND DISCUSSION
Studying genetic diversity for traits and using this diversity for genetic improvement of traits is of paramount importance.
Investigation of plant height in sesame genotypes: Analysis of variance and the height diagram of the stem, genetic Variance was 1785.16 cm and phenotype variance has the lowest height of 1962.26 cm (Table 1 and 2). These results agreed with Bhuiyan et al.2 which shows variance and the height diagram of the stem and Naz single-branched cultivar has the highest height and the local Mehran cultivar.
Examining the number of buds in sesame genotypes: According to the variance analysis table for the number of buds, Nashkofa variety has the highest average number of buds 205.87 and Dashtestan2 has the lowest number of buds 0.744 (Table 2). Chowdhury et al.3 have found similar results that sesame genotypes affected with a number of buds.
Examining the percentage of chlorophyll in sesame genotypes: According to variance analysis table for the percentage of chlorophyll, the highest average chlorophyll percentage is 118. 93 and the lowest chlorophyll percentage is 82.13 (Table 2-4). Kadvani et al.5 have indicated that in variance analysis for the percentage of chlorophyll, Naz variety has the highest average chlorophyll percentage and Mehran local variety.
Examining the number of seeds in sesame genotypes: Table (2 and 5) show that genetic variance has been recorded at 71.44 and phenotype variance was 97.13. These results were agreed with previous studies6,12-14.
Investigation of plant performance in sesame genotypes: According to the variance analysis table, yellow white variety has the highest average plant yield and Dashtestan variety has the lowest plant yield (Table 6).
| Table 1: | Variance analysis for plant height (cm) using SS adjusted for testing | |||
| Resources of changes | Degree of freedom | Average of squares | F |
| Block | 2 | 4710.3 | 26.6 |
| Cultivar | 15 | 5178.4 | 29.24 |
| Test error | 30 | 3383.6 | 19.1 |
| Sample error | 192 | 177.1 | |
| Total | 239 | ||
| Grouping cultivars based on plant height using Tukey's method and 95.0% confidence | |||
| Table 2: | Variance analysis for the number of buds using SS adjusted for the experiment | |||
| Variance components | Heritability | |||
| Traits | Genetic variance | General | Phenotype variance | General |
| Diameter of stem | 0.33 | 0.892 | 0.37 | 0.892 |
| Plant height | 1785.16 | 0.91 | 1962.26 | 0.91 |
| Number of buds | 205.87 | 0.744 | 276.64 | 0.744 |
| Percent of chlorophyll | 82.13 | 0.691 | 118.93 | 0.691 |
| Number of branch | 0.519 | 0.499 | 1.04 | 0.499 |
| Number of seed | 71.44 | 0.736 | 97.13 | 0.736 |
| Whole plant performance | 0.17 | 0.0103 | 0.18 | 0.944 |
| Plant yield | 1.12 | 0.067 | 1.18 | 0.949 |
| Grouping cultivars based on the number of buds using Tukey's method and 95.0% confidence | ||||
| Table 3: | Variance analysis table for chlorophyll percentage using SS adjusted for experiment | |||
| Resources of changes | Degree of freedom | Average of squares | F |
| Block | 2 | 4.5 | 0.03 |
| Cultivar | 15 | 209.6 | 1.25 |
| Test error | 30 | 36.8 | 0.22 |
| Sample error | 192 | 167.8 | |
| Total | 239 | ||
| Grouping cultivars based on chlorophyll percentage using Tukey’s method and 95.0% confidence | |||
| Table 4: | Diagram of chlorophyll percentage in studied genotypes | |||
| Resources of changes | Degree of freedom | Average of squares | F |
| Block | 2 | 5.85 | 11.23 |
| Cultivar | 15 | 1.0375 | 1.99 |
| Test error | 30 | 0.9567 | 1.84 |
| Sample error | 192 | 0.5208 | |
| Total | 239 | ||
| Grouping cultivars based on number of branches using the Tukey’s method and 95.0% confidence | |||
| Table 5: | Variance analysis table for seed count using SS adjusted for testing | |||
| Resources of changes | Degree of freedom | Average of squares | F |
| Block | 2 | 380.71 | 14.83 |
| Cultivar | 15 | 188.71 | 7.35 |
| Test error | 30 | 219.31 | 8.54 |
| Sample error | 192 | 25.67 | |
| Total | 239 | ||
| Grouping cultivars based on seed number using Tukey's method and 95.0% confidence | |||
| Table 6: | Variance analysis for plant performance using SS adjusted for testing | |||
| Resources of changes | Degree of freedom | Average of squares | F |
| Block | 2 | 0.0308 | 0.45 |
| Cultivar | 15 | 3.2972 | 48.61 |
| Test error | 30 | 0.0239 | 0.35 |
| Sample error | 192 | 0.0678 | |
| Total | 239 | ||
| Grouping cultivars based on plant performance using Tukey's method and 95.0% confidence | |||
| Table 7: | Analysis of variance for whole plant performance using SS adjusted for testing | |||
| Resources of changes | Degree of freedom | Average of squares | F |
| Block | 2 | 0.00469 | 0.45 |
| Cultivar | 15 | 0.50151 | 48.61 |
| Test error | 30 | 0.00364 | 0.35 |
| Sample error | 192 | 0.01032 | |
| Total | 239 | ||
| Grouping of cultivars based on whole plant yield using Tukey's method and 95.0% confidence | |||
Examining the performance of the whole plant in sesame genotypes: According to variance analysis and general heritability of traits, genetic variance was recorded at 0.17 and phenotype variance was recorded at 0.18 has the lowest yield of the whole plant (Table 2 and 7).
CONCLUSION
The study concludes that the heritability of all traits was high, except for day to maturity, which had moderate heritability. The General Average (GA) was high for all traits except weight of 1000 seeds, days to 50% flowering, capsule length, protein content, oil content and days to maturity. The evaluating seed yield of four cultivars named Mehando-80, T, Hummera-1 and in Southern Region of Ethiopia showed that the highest seed yield related to 80-Mehando cultivar and the lowest seed yield related to 85-T cultivar in the studied area.
SIGNIFICANCE STATEMENT
Performance is a complex trait influenced by various factors that are inherited in a polygenic manner and highly subject to environmental changes. Therefore, functional components that are less affected by the environment should be selected. The aim of this study was to evaluate of genetic parameters of seed yield in sesame cultivars. The study of correlation between performance and its components and their relative contribution to performance in breeding programs is of great importance. The results showed that the heritability of all traits was high, except for day to maturity, which had moderate heritability. The General Average (GA) was high for all traits except weight of 1,000 seeds, days to 50% flowering, capsule length, protein content, oil content and days to maturity.
ACKNOWLEDGMENT
The authors would like to thank Ilam University, college of Agriculture, Iran and Wasit University/ college of Agriculture, Department of Crop Sciences, Iraq for their support.
REFERENCES
- Aboelkassem, K.M., A.A. Ahmed and T.H.A. Hassan, 2021. Genetic variability and path coefficient analysis of some sesame (Sesamum indicum L.) genotypes for seed yield and their components. J. Plant Prod., 12: 377-382.
- Bhuiyan, M.S.H., M.A. Malek, M.A. Sarkar, Majharul Islam and W. Akram, 2019. Genetic variance and performance of sesame mutants for yield contributing characters. Malays. J. Sustainable Agric., 3: 27-30.
- Chowdhury, S., A.K. Datta, A. Saha, S. Sengupta, R. Paul, S. Maity and A. Das, 2010. Traits influencing yield in sesame (Sesamum indicum L.) and multilocational trials of yield parameters in some desirable plant types. Indian J. Sci. Technol., 3: 163-166.
- Ebrahimian, E., S.M. Seyyedi, A. Bybordi and C.A. Damalas, 2019. Seed yield and oil quality of sunflower, safflower, and sesame under different levels of irrigation water availability. Agric. Water Manage., 218: 149-157.
- Kadvani, G., J.A. Patel, J.R. Patel, K.P. Prajapati and P.J. Patel, 2020. Estimation of genetic variability, heritability and genetic advance for seed yield and its attributes in sesame (Sesamum indicum L.). Int. J. Bio-Resour. Stress Manage., 11: 219-224.
- Kumar, S., V. Kumari and V. Kumar, 2018. Genetic variability and character association studies for seed yield and component characters in soybean [Glycine max (L.) Merrill] under North-Western Himalayas. Legume Res., 43: 507-511.
- Monpara, B.A. and S.S. Khairnar, 2016. Heritability and predicted gain from selection in components of crop duration and seed yield in sesame (Sesamum indicum L.). Plant Gene Trait, 7.
- Mansouri, S., M. Esmailov and M.A. Sarbarzeh, 2016. Evaluation of genetic parameters and combining ability of important agronomic traits in sesame using diallel cross. J. Seedling Seed Breed., 32: 119-140.
- Bazaz, A.M., A. Tehranifar, M. Kafi, A. Gazanchian and M. Shoor, 2015. Effect of salinity on seed germination and seedling growth of native populations of tall fescue in Iran. J. Hort. Sci., 29: 269-276.
- Muhammad, L.M., O.A. Falusi, O.A.Y. Daudu, A.A. Gado and S.A. Yahaya, 2013. Radiation induced polygenic mutation in two common Nigerian sesame (Sesamum indicum L.) cultivars. Int. J. Biotechnol. Food Sci., 1: 23-28.
- Teimouri, M.S., H.R. Khazaie, M.N. Mahallati and A. Nezami, 2010. Effect of salinity on seed yield and yield components of individual plants, morphological characteristics and leaf chlorophyll content of sesame (Sesamum indicum L.). Environ. Stresses Agric. Sci., 2: 119-130.
- Azeem, M., M. Qasim, M.W. Abbasi, Tayyab, R. Sultana, M.Y. Adnan and H. Ali, 2019. Salicylic acid seed priming modulates some biochemical parameters to improve germination and seedling growth of salt stressed wheat (Triticum aestivum L.). Pak. J. Bot., 51: 385-391.
- Pratap, A., P. Bisen, B. Loitongbam, Sandhya and P.K. Singh, 2018. Assessment of genetic variability for yield and yield components in rice (Oryza sativa L.) germplasms. Int. J. Bio-Resour. Stress Manage., 9: 87-92.
- Radhakrishnan, R. and I.J. Lee, 2015. Penicillium-sesame interactions: A remedy for mitigating high salinity stress effects on primary and defense metabolites in plants. Environ. Exp. Bot., 116: 47-60.
How to Cite this paper?
APA-7 Style
Ibrahim,
A.J., Tahmasbe,
Z.J. (2024). Evaluation of Genotype Coefficient of Variation (GCV) and Phenotypic Coefficient of Variation (PCV) of Seed Yield in Sesamum indicum Cultivars. Asian Journal of Biological Sciences, 17(4), 567-573. https://doi.org/10.3923/ajbs.2024.567.573
ACS Style
Ibrahim,
A.J.; Tahmasbe,
Z.J. Evaluation of Genotype Coefficient of Variation (GCV) and Phenotypic Coefficient of Variation (PCV) of Seed Yield in Sesamum indicum Cultivars. Asian J. Biol. Sci 2024, 17, 567-573. https://doi.org/10.3923/ajbs.2024.567.573
AMA Style
Ibrahim
AJ, Tahmasbe
ZJ. Evaluation of Genotype Coefficient of Variation (GCV) and Phenotypic Coefficient of Variation (PCV) of Seed Yield in Sesamum indicum Cultivars. Asian Journal of Biological Sciences. 2024; 17(4): 567-573. https://doi.org/10.3923/ajbs.2024.567.573
Chicago/Turabian Style
Ibrahim, Azhar, Jabbar, and Zahra Jabbar Tahmasbe.
2024. "Evaluation of Genotype Coefficient of Variation (GCV) and Phenotypic Coefficient of Variation (PCV) of Seed Yield in Sesamum indicum Cultivars" Asian Journal of Biological Sciences 17, no. 4: 567-573. https://doi.org/10.3923/ajbs.2024.567.573

This work is licensed under a Creative Commons Attribution 4.0 International License.

