Rapid Screening Methods for Identification of Resistant Maize Inbreds to Fungal Foliar Pathogens
| Received 22 Jul, 2024 |
Accepted 10 Sep, 2024 |
Published 31 Dec, 2024 |
Background and Objective: Maize (Zea mays L.) is an important cereal that provides nutrition for about 50% of the global population. Despite the numerous benefits and potential of the crop, maize is susceptible to various biotic stresses that limit its maximum yield potential. Host plant resistance (HPR) has been adjudged the best option among the various options for disease management. However, identifying sources of resistance among maize genotypes maximizes the efficiency of HPR. The present study compared the effectiveness of the rapid laboratory assay; the detached leaf assay (DLA) with screen house screening for rapid identification of disease-resistant maize genotypes. Materials and Methods: Ten each of the early and extra-early maturing maize inbreds were screened for resistance to Bipolaris maydis (SLB 02) and Exserohilum turcicum (NGIB16-13), respectively. The screen house experiment was laid in a restricted randomized complete block design with two replications and disease severity was measured using a 1-9 scale weekly after inoculation. Four-weeks-old leaves of each genotype were inoculated with 40 µL of 10-5 spores/mL of the pathogens using the DLA, in a completely randomized design. Data collected were subjected to ANOVA and significant means were separated using Fisher’s LSD at 0.05 probability level. Results: The DLA and screen house experiments were compared and were found to be similar. The TZEEI 179, TZEEI 8 and TZEEI 20 were identified as SCLB-resistant inbreds while TZEI 56, TZEI 60, TZEI 86 and TZEI 128 were found to be NCLB-resistant genotypes using both methods. Conclusion: The DLA is an efficient technique for identifying disease-resistant early and extra-early maturing genotypes and could facilitate the rapid development of disease-resistant maize hybrids and open-pollinated varieties. Adopting the disease-resistant genotypes would reduce crop losses, increase farmer’s income and ensure food security.
| Copyright © 2024 Bankole et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Plants are important for the survival of life. Besides providing food for humans and other animals, plants also support a wide variety of microbes that are part of the ecosystem1. Despite man’s efforts to use plants for his benefit, competing microbes continue to defy him and take a significant portion of the resources that man would like to exploit for his use. As a result, diseases develop and spread, which lowers plant production and yield1.
Maize (Zea mays L.) is the primary staple food crop widely cultivated across sub-Saharan Africa (SSA)2 and is farmed in rainfed and irrigated areas. It is called the “Queen of Cereals” due to its widespread popularity and higher potential than any other cereal3,4. It is an energy-rich resource with 65% starch, 12% protein and 6% oil, along with husk and bran (fibre). Maize accounts for 15% of the world’s protein and supplies 19% of the calories obtained from food crops5. Maize has great potential for production in West and Central Africa (WCA) and yields more than other cereal crops6. More than 300 million Africans depend on it as their primary staple meal, making it the most significant food in SSA and thus essential to food security7.
Maize belongs to the kingdom; Plantae, Class; Tracheophytes, Clade; Angiosperms, order; Poales, Family; Poaceae, Subfamily; Panicodeae, Genus, Zea and species; mays8. Zea, an ancient Greek term that means "supporting life" and mays, a Taino word that means "life provider" are both related concepts. The crop has fulfilled its name as it is a life-saving crop used in several ways9,10. The economic relevance of maize extends across various areas of human existence since it functions as food for humans, such as pap, thick porridge, popcorn and boiling grains, which are popular foods enjoyed by the majority of Nigerians, particularly in the country's southern region. Maize is required for the manufacture of starch and alcohol. The starch can be utilized as a dextrin, sugar and syrup converter; maize oil can be used to produce soup or refined for cooking and salad dressing. Maize extracts, particularly corn silk, have been used to treat urinary system disorders and kidney-related illnesses, while different portions of the crop, such as the root, are used to cure abdominal abnormalities11,12.
Maize was domesticated in Southern Mexico/Meso America over 9000 years ago13. Since its domestication, maize has spread rapidly worldwide and since become a primary staple cereal globally with annual production surpassing 1 billion metric tons14. In 2021, the total world production of maize was 1.16 million tons with America, Asia, Europe and Africa, producing 49, 34, 9 and 8%, respectively, of total world production2.
For both basic and practical maize research, inbred lines have proven to be great resources. They represent a sample of maize genetic diversity, which has been recorded and divided into a variety of consistent, reproducible genotypes10. For example, extra early-maturing maize (EEM genotypes that reach physiological maturity between 80 to 85 days) and early-maturing maize (EM genotypes mature physiologically between 90 and 95 days) were developed and utilized for the development of various maize genotypes with improved qualities such as Striga hermonthica and Exserohilum turcicum resistance, tolerance to drought, heat and low-soil-nitrogen as well as nutrition qualities; provitamin A and quality protein15.
Despite the numerous benefits and potential of the crop, maize is susceptible to biotic and abiotic stresses that limit the maximum yield potential of the crop15,16. The biotic stresses include diseases caused by nematodes, viruses, bacteria and fungi, while drought, low soil fertility and heat stress are the abiotic factors that limits maize productivity and results in low quality and crop yield17. Of the biotic agents, fungal infections significantly reduce maize production and productivity18.
Fungi are achlorophyllous eukaryotic, spore-forming organisms, the majority of which are filamentous and branched, with over 100,000 species as saprophytes. However, more than 20,000 species are phytopathogenic, causing diseases in crops and plants19. Phytopathogenic fungi play significant roles in annual crop yield losses and are consequently, of significant economic concerns for farmers and scientists20,21. Defoliation, chlorotic and necrotic leaf lesions caused by fungal infections decrease the photosynthetic potential of the crop, resulting in reduced productivity. Some destructive maize foliar diseases have been reported which include: Southern corn leaf blight (SCLB), incited by Bipolaris maydis
(Nisik. Y. and Miyake C.) Shoemaker, northern corn leaf blight (NCLB), by Exserohilum turcicum (Pass.) Leonard and Suggs; gray leaf spot, incited by Cercospora zeae-maydis Tehon and Daniels22 and curvularia leaf spot, caused by Curvularia lunata (Wakker) Boedijn15,23. Among these, SCLB and NCLB are prominent in maize fields in Nigeria16,24-26.
Bipolaris maydis is a hemibiotrophic ascomycete previously called Helminthosporium maydis (Teleomorph; Cochliobolus heterostrophus)8. Bipolaris maydis was first discovered in the United States in 1923. Between the years 1970-1971, it reached pandemic proportions, decimating 15% of the maize crop in the United States and Southern Canada for US$1.0 billion (about $6.0 billion by 2015 standards)8. Presently, the pathogen affects tropical and subtropical maize-growing regions in the Southeast United States, as well as some regions of Asia and Africa25,26. This fungus' spores fall, germinate on the leaf surface and could either pass through the stomata or leaf cuticle and epidermis27. Within days, this pathogen can create windborne spores that spread through leaf litter28. Infected leaves develop distinctive lesions as the fungal infection develops and the allocation of resources to reproductive tissues decreases27,29.
Another devastating pathogen of serious concern in the tropical lowlands of Nigeria where maize is grown is Exserohilum turcicum causing NCLB. The disease can cause between 30 - 70% grain yield loss depending on host resistance, disease severity, plant's growth stage during infection and position of the infected leaves7,10,30,31. High disease severity during the reproductive stages of the crop, results in severe yield losses that range between 40 and 70%16.
Various management techniques, including disease-resistant cultivars, crop rotation, seed treatment and application of fungicides, have been employed to mitigate crop losses. However, host plant resistance (HPR) has been adjudged an appropriate, affordable, sustainable and environmentally friendly management option15,32,33. Plant genetic resistance is classified into two: Qualitative and quantitative resistance. Qualitative resistance is based on a single major-effect resistance gene (R gene) offering race-specific, high-level resistance, while quantitative resistance is frequently multigenic and provides non-race-specific intermediate degrees of resistance. Qualitative resistance, when deployed, is frequently linked with a fast cell death known as a hypersensitive response (HR) surrounding the area of pathogen entry and is largely easy to breakdown, whereas, quantitative resistance lasts longer34. However, for the effective development of disease-resistant varieties, identification of genotypes with resistance becomes important. Scientists have often used field screening methods for the identification of resistant genotypes because it provides information on the plant-pathogen interactions and facilitates accurate identification of resistant genotypes. However, the method is labor-intensive and requires a lengthy period of time especially when a large number of genotypes are to be screened. This necessitates the need for a rapid screening method that could discriminate genotypes to a variety of foliar pathogens. Detached leaf assay (DLA) facilitates screening of several genotypes under controlled environments and promotes homogeneous disease development with standard inoculum loads15. It is a quick, inexpensive laboratory-based method for identifying genotypes with resistance to B. maydis and E. turcicum15,26,35,36. In an integrated disease management approach, host plant resistance has been adjudged the best option, it is therefore, important to investigate a rapid and accurate methods for identifying disease-resistant genotypes. The present study evaluated the effectiveness of the rapid laboratory assay; the detached leaf assay (DLA) in comparison with screenhouse experiments for identifying disease-resistant maize genotypes.
MATERIALS AND METHODS
Experimental location: The experiments were carried out at the Microbiology Laboratory and Screenhouse Facility of the Department of Microbiology and Biotechnology, Faculty of Natural and Applied Sciences, First Technical University, Ibadan, Nigeria (7°14'N, 3°49'E). The study was conducted during the 2022 growing season between March, 2022 to November, 2022.
Genotype composition and evaluated fungi: Ten each of EM and EEM maize inbreds (consisting 5 white and 5 yellow endosperm each) were obtained from the International Institute of Tropical Agriculture (IITA) Ibadan, Nigeria.
Screenhouse experiment: Sterile topsoil was dispensed into 12 L pots. Four seeds each of EM and EEM maize inbreds were sown per pot. Following emergence, the plants were thinned to two plants per pot, 2 weeks after planting. Basal application of NPK 15-15-15 fertilizer was done 3 weeks after planting (WAP) and top-dressed with urea five WAP.
Preparation of growth media: Potato dextrose agar was prepared according to manufacturer’s instruction. Briefly, 39 g of PDA was added to 1000 mL of distilled water, it was homogenized using the magnetic stirrer for five minutes and sterilized using the autoclave at 121°C for 15 min. It was left to cool, lactic acid (1 mL/L) and streptomycin (0.3 g/L) was added to adjust its pH and inhibit bacteria growth, respectively. Afterwards, the media was dispensed into sterile petri dishes aseptically in a class ІІ biosafety cabinet and left to solidify. In addition, V8 (20% vegetable juice) agar medium was prepared by adding 200 mL of V8 juice to 800 mL of distilled water. Afterwards, 3 g/L of CaCO3 and 20 g/L of agar-agar powder, respectively, were added. The suspension was homogenized and sterilized as discussed earlier. The suspension was left to cool and streptomycin (0.3 g/L) and neomycin (0.05 g/L) were added to inhibit the growth of bacteria. The media was afterwards poured into sterile petri-dishes aseptically in a class ІІ biosafety cabinet and left to solidify.
Inoculum preparation for screenhouse studies: Following the protocol established of Badu-Apraku et al.7and Bankole et al.15, 100 g of white sorghum was measured into 250 mL Erlenmeyer flasks and soaked overnight. After 24 hrs, the water was decanted, grains washed and the flasks were covered with aluminum foil and sterilized in an autoclave at 121°C for 1 hr. Conidia from 14-day-old cultures of B. maydis (SLB 02) and E. turcicum (NGIB16-13), were harvested with the aid of sterile plastic spreader and Tween-20 solution and adjusted to 105 spores/mL using a Neubauer haemocytometer (CorningTM, UK). The flasks containing sterilized sorghum grains were inoculated aseptically with 4 mL of the spore suspension7. The inoculated flasks were thoroughly shaken to ensure uniform spread of the spore suspension. The inoculated flasks were incubated at room temperature for seven days. The flasks were shaken daily for even colonization by the inoculum. After 7 days of inoculation and incubation, the pathogens sporulated and colonized the sterilized sorghum.
Screenhouse inoculation: The whorl of 28-day-old maize plants was inoculated with 20 g of B. maydis (SLB 02) and E. turcicum (NGIB16-13) colonized sorghum grains independently. The EEM and EM inbred lines were individually challenged with B. maydis (SLB 02) and E. turcicum (NGIB16-13). A restricted randomized complete block design with two replications was used for the screen house experiment. The restriction was based on the different pathogens (B. maydis and E. turcicum) used.
Disease assessment: Plants were visually assessed for disease severity from one week after inoculation up to six weeks after inoculation (DS1WAI-disease severity 1 week after inoculation, DS2WAI, DS3WAI, DS4WAI, DS5WAI and DS6WAI) using the scale of 1-97. A field book application by Rife and Poland37 was used for data collection.
Measurement of agronomic traits: The number of plants per pot, the day when 50% of the plants in a pot shed pollen and the number of days from planting to anthesis (DA) or incipient silk (DS) extrusion were all recorded. The anthesis-silking interval (ASI) was calculated as the difference between DS and DA. Plant height (PLHT) and ear height (EHT) were computed as the distance from the plant’s base to the height of the first-tassel branch and, accordingly, to the node containing the upper ear. Plant aspect (PASP) was assessed on a scale of 1-9, with 1 representing high phenotypic appeal and 9 representing
poor phenotypic appeal, based on variables such as relative uniformity of plant and ear placement, uniformity, sensitivity to diseases and insects and lodging. Ear aspect (EASP) considered the severity of disease and insect damage visible in ears, as well as ear size and uniformity. The EASP was graded on a scale of 1-9, with 1 representing clean, uniform, broad and well-filled ears and 9 representing ears with undesirable traits. The rating scales used for PASP, EASP was adapted from Badu-Apraku et al.7, the weight of the grains (GWT) was obtained using the digital weighing balance (Yoke instrument, Shanghai, China).
Detached leaf assay (DLA): Crisper boxes were sterilized by soaking in 20% Sodium hypochlorite (NaOCl) and mancozeb solution for 2 min in a biosafety cabinet (Class II). Technical agar (TA; 10 g/L) amended with 6-benzylaminopurine phytohormone (0.045 g/L) was autoclaved at 121°C for 15 min. After cooling, lactic acid (1.5 mL/L) and hexaconazole (8.33 mg/L) were added to prevent contamination. Afterwards, 250 mL of TA was dispensed into each sterile box and allowed to set within the biosafety cabinet15,26,35.
Pathogenicity test using DLA: Each inbred’s fully developed leaves were carefully selected, harvested and immediately placed in moistened polythene bags and transferred to the laboratory. The leaves were cut into 6×3 cm pieces and surface sterilized for 90 sec in 1% NaOCl solution. Afterwards, the leaves were rinsed in three changes of sterile distilled water. Excess water was blotted with sterile paper towels in the biosafety cabinet. The harvested leaves were carefully placed in the sterile crisper boxes containing the TA medium described above. The adaxial side of the leaves were placed on the medium15,35. Each leaf section was inoculated individually with 40 μL of spore suspensions of SLB 02 and NGIB-16-13, independently. Sterile water was used to inoculate the control treatments. Boxes were covered and sealed with cling film before being transferred to the laboratory bench tops for incubation at room temperature (about 25°C) and a 12 hrs photoperiod. The inoculated leaves were inspected at 2-days intervals for symptom manifestation for 10 days. Disease severity was determined using a scale of 1-5, with ‘1 indicating no obvious symptoms, 2 indicating 1-10% leaf area covered with brownish chlorosis, 3 indicating 11-25% leaf area covered with brownish lesions, 4 indicating 26-50% leaf area covered with brownish lesions and 5 indicating >50% leaf area covered with brownish lesions15.
Pathogenicity test using Koch’s postulate: Infected leaves were excised, rinsed to get rid of weevils and taken to the laboratory. Using a scalpel blade, the leaves were sectioned into small pieces including ¼ of the infected area and ¾ of uninfected area. The leaf segments were surface sterilized using 70% ethanol, 20% NaOCl solution and properly rinsed with sterile distilled water in the biosafety cabinet. Sterile paper towels were used to blot excess water from the segment. Afterwards, the segments were plated on PDA plates and incubated at 28°C for 3 days and fungal growth was observed. Each fungal growth noticed on the media was sub-cultured until pure cultures of each isolate was obtained. The recovered isolates were examined under the microscope (Olympus BX53, Japan, Magnification ×40) to confirm Koch’s postulate.
Data analysis: The collected data on agronomic traits (plant height (cm), ear height (cm), plant aspect, ear aspect, grain yield (g) etc.) and disease severity were subjected to analysis of variance employing a fixed model using Statistical Analysis Software (SAS®) version 9.4. For each of the maize genotype, significant means were separated using the Fisher’s least significant difference (LSD) at 0.05 level of probability.
RESULTS
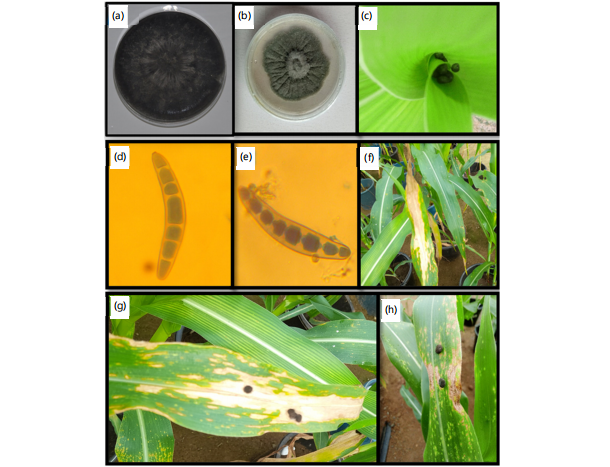
Early maturing inbred lines were independently inoculated with B. maydis (SLB 02) and E. turcicum (NGIB16-13). The isolates of each pathogen were pathogenic following Koch’s postulate. The reaction of TZEEI 157 to B. maydis isolate (SLB 02) and TZEI 1 to E. turcicum (NGIB16-13) is presented in Fig. 1(a-h).
|
Additionally, the maize genotypes reacted differently (p<0.05) to the inoculated pathogens based on measured agronomic traits and on disease severity. Furthermore, both the DLA and screenhouse evaluations were adequate for the identification of resistant inbred lines. The average values of the agronomic data obtained from extra-early maize inbreds inoculated with B. maydis are presented in Table 1. The TZEEI 86 had better agronomic features compared to other inbreds. Following inoculation with B. maydis, isolate SLB 02, the inbred (TZEEI 86) had lowest plant aspect (PASP) and ear aspect (EASP) which indicated good overall plant phenotypic appeal and very good ear aspect, respectively. The cob length measured as ear height (19.2 cm) indicated that the inbred had long cobs which implied accumulation of more grains compared to inbreds with shorter cob length. In addition, the inbred had the highest grain weight (42.2 g). The TZEEI 157 had PASP and EASP values of 4 (satisfactory plant aspect) and 3 (good ear aspect), respectively and a grain weight of 32 g. Furthermore, TZEEI, 79 and TZEEI 179, had moderate plant and ear aspects rating scores (4), shorter cob length and lower grain weight compared to TZEEI 86. Although, TZEEI 158 had similar plant aspect and ear aspect ratings with TZEEI 79 and TZEEI 179, the grain weight of TZEEI 158 was higher (p<0.05) than either TZEEI 79 and TZEEI 179. The TZEEI 20, had an acceptable plant aspect rating, but a good ear aspect rating that culminated in grain weight of 28.3 g. However, TZEEI 51 had the most undesirable agronomic traits, with a relatively acceptable PASP but an undesirable EASP score and grain weight.
| Table 1: | Average values of selected agronomic traits of inbred lines inoculated with Bipolaris maydis (SLB 02) | |||
| Genotype | Plant height (cm) | Ear height (cm) | Plant aspect** | Ear aspect** | Grain weight** (g) |
| TZEEI 1 | 124 | 17.1 | 5 | 5 | 22.3 |
| TZEEI 157 | 125 | 22.3 | 4 | 3 | 32.3 |
| TZEEI 158 | 140 | 19.2 | 4 | 4 | 30.1 |
| TZEEI 179 | 67 | 11.4 | 4 | 4 | 18.4 |
| TZEEI 20 | 139 | 18.2 | 5 | 4 | 28.3 |
| TZEEI 32 | 68 | 7.4 | 5 | 4 | 16.1 |
| TZEEI 51 | 135 | 14.1 | 5 | 7 | 10.4 |
| TZEEI 79 | 161 | 15.9 | 4 | 4 | 14.2 |
| TZEEI 8 | 142 | 17.1 | 5 | 5 | 18.3 |
| TZEEI 86 | 159 | 19.2 | 3 | 4 | 42.2 |
| LSD (0.05) | 105 | 5.1 | 2 | 5 | 11.3 |
| LSD: Least significant difference and **Connotes traits with statistical significance at p<0.01 | |||||
| Table 2: | Average values of the selected agronomic traits of inbred lines inoculated with Exserohilum turcicum (NGIB-16-13) | |||
| Genotype | Plant height (cm) | Ear height (cm) | Plant aspect** | Ear aspect** | Grain weight** (g) |
| TZEI 1 | 159.5 | 15.8 | 3 | 5 | 17.9 |
| TZEI 120 | 113 | 14 | 4 | 5 | 15.2 |
| TZEI 124 | 167 | 16.5 | 3 | 4 | 16 |
| TZEI 128 | 136 | 16 | 5 | 5 | 21.5 |
| TZEI 134 | 133 | 18.7 | 6 | 4 | 16 |
| TZEI 135 | 154.5 | 15 | 4 | 4 | 28 |
| TZEI 269 | 120.5 | 17.5 | 4 | 4 | 20.5 |
| TZEI 56 | 137.5 | 12.7 | 5 | 4 | 22.6 |
| TZEI 60 | 180 | 17.5 | 4 | 3 | 32 |
| TZEI 86 | 124 | 16 | 4 | 3 | 30 |
| LSD (0.05) | 17.4 | 8.9 | 1 | 1 | 4.5 |
| LSD: Least significant difference and **Connotes traits with statistical significance at p<0.01 | |||||
The effect of Exserohilum turcicum, isolate NGIK-16-13 on the agronomic traits of early maturing maize inbred lines is presented in Table 2. The TZEI 60 had the highest plant height of 180 cm, followed by TZEI 124 with a plant height of 167 cm, TZEI 1 had a plant height of 159.5 cm and TZEI 120 had the lowest plant height of 113 cm. For plant aspect ratings, TZEI 1 and TZEI 124 had the lowest PASP score of 3 which indicates good phenotypic appeal under E. turcicum inoculation. The TZEI 60, TZEI 86, TZEI 120, TZEI 135 and TZEI 269, had satisfactory phenotypic appeal. However, the highest plant aspect rating of 6 indicating undesirable phenotypic appeal, was recorded from TZEI 134. In addition, TZEI 60 and TZEI 86 had the lowest EASP score of 3 which is considered to be the best ear aspect rating, followed by TZEI 134, TZEI 135, TZEI 269, TZEI 56 and TZEI 124 with satisfactory ear aspect ratings. However, TZEI 1, TZEI 120 and TZEI 128 had the highest ear aspect rating. The highest grain weight was recorded from TZEI 60, TZEI 86 and TZEI 135 with grain weights of 32, 30 and 28 g, respectively. The TZEI 86 and TZEI 60, combined good agronomic traits with grain weight. Contrarily, TZEI 124 with good phenotypic appeal had reduced grain weight in the present study.
The average disease severity (DS) scores of extra-early maize inbred lines challenged with B. maydis (SLB 02) are presented in Table 3. Under screenhouse conditions, there was no significant difference (p>0.05) in the reactions of the various genotypes to the pathogen until the 4th week after inoculation (WAI). The disease severity score 4WAI revealed variation (p<0.05) in the response of the inbred to the pathogen. The TZEEI 8 had the least disease severity score followed by TZEEI 179. The disease severity scores indicated resistance to the pathogen. The TZEEI 20, TZEEI 79, TZEEI 1, TZEEI 51 and TZEEI 86 were moderately resistant to the pathogen. However, TZEEI 158 and TZEEI 32 were found to be susceptible to the pathogen. For DS5WAI, although the DS scores increased for the different inbred lines, the reaction patterns of the inbred lines were similar. At 6WAI, i.e. 10 weeks after planting, TZEEI 179 had the lowest disease severity score while TZEEI 158 and TZEEI 32 had the highest disease severity score and were susceptible to the pathogen. Evaluating four-weeks-old leaves of the inbred lines for resistance using the detached leaf assay (DLA), the inbred lines responded similarly to those evaluated under the screenhouse conditions. For example, at 10 days after inoculation (DAI), TZEEI 179 was found to be the most resistant inbred with DS score of 3.0, while TZEEI 158 and TZEEI 32 were found to be susceptible (DS score = 5 and Table 3). These results are consistent with those obtained for the same inbred lines evaluated under the screenhouse conditions.
| Table 3: | Disease severity scores of extra-early maize inbreds inoculated with B. maydis under screenhouse conditions and detached leaf assay | |||
| Screenhouse | Detached leaf assay | |||||
| Genotype | DS4WAI* | DS5WAI* | DS6WAI* | DS6DAI* | DS8DAI* | DS10DAI* |
| TZEEI 179 | 1.8 | 1.9 | 2.6 | 1.8 | 1.8 | 3.0 |
| TZEEI 8 | 1.6 | 2.4 | 3.3 | 2.5 | 3.3 | 3.6 |
| TZEEI 20 | 2.5 | 3.4 | 3.8 | 2.8 | 3.5 | 3.8 |
| TZEEI 79 | 2.6 | 3.4 | 4.5 | 3.0 | 3.3 | 4.0 |
| TZEEI 1 | 3.0 | 5.0 | 5.0 | 3.6 | 4.2 | 4.6 |
| TZEEI 51 | 3.7 | 4.2 | 5.3 | 3.6 | 4.5 | 4.8 |
| TZEEI 86 | 3.6 | 4.5 | 5.4 | 3.8 | 4.6 | 5.0 |
| TZEEI 157 | 4.8 | 5.5 | 6.0 | 3.4 | 5.0 | 5.0 |
| TZEEI 158 | 6.4 | 6.8 | 7.4 | 4.4 | 5.0 | 5.0 |
| TZEEI 32 | 6.5 | 7.5 | 7.8 | 4.6 | 5.0 | 5.0 |
| LSD (0.05) | 1.2 | 2.0 | 2.2 | 0.4 | 0.8 | 1.2 |
| LSD: Least significant difference, *Connotes traits with statistical significance at p<0.05 and Bipolaris maydis SLB 02 was used for inoculation | ||||||
| Table 4: | Disease severity scores of early maize inbreds inoculated with Exserohilum turcicum under screenhouse conditions and detached leaf assay | |||
| Screenhouse | Detached leaf assay | |||||
| Genotype | DS4WAI* | DS5WAI* | DS6WAI* | DS6DAI* | DS8DAI* | DS10DAI* |
| TZEI 60 | 1.8 | 3.7 | 4.4 | 1.8 | 2.2 | 3.0 |
| TZEI 56 | 3.0 | 4.2 | 4.5 | 1.6 | 2.0 | 2.7 |
| TZEI 128 | 3.4 | 3.8 | 4.2 | 2.0 | 2.5 | 3.0 |
| TZEI 86 | 3.2 | 4.6 | 5.0 | 1.8 | 2.4 | 2.7 |
| TZEI 134 | 3.0 | 4.8 | 4.8 | 2.0 | 2.5 | 3.7 |
| TZEI 124 | 3.0 | 4.6 | 5.4 | 2.0 | 2.8 | 4.0 |
| TZEI 269 | 3.2 | 4.8 | 5.8 | 2.8 | 3.0 | 3.0 |
| TZEI 120 | 3.4 | 4.6 | 5.8 | 3.0 | 3.5 | 3.7 |
| TZEI 135 | 3.6 | 4.8 | 6.0 | 3.0 | 3.3 | 4.0 |
| TZEI 1 | 3.5 | 4.8 | 6.4 | 3.0 | 3.3 | 4.3 |
| LSD (0.05) | 0.6 | 0.4 | 0.4 | 0.2 | 0.5 | 0.3 |
| LSD: Least significant difference, *Connotes traits with statistical significance at p<0.05 and Exserohilum turcicum isolate NGIB16-13 was used for inoculation | ||||||
The reaction of early maturing inbred lines to Exserohilum turcicum isolate NGIB-16-13 evaluated under the screenhouse conditions are presented in Table 4. The 4WAI and TZEI 60 had the least DS score (1.8) which was significantly different from others inbreds. Additionally, other inbreds were not severely infected by the pathogen at 4WAI. At 5WAI, the reaction of the different inbred lines was noticeable as the inbred lines varied significantly in their reaction to the pathogen. For example, TZEI 60, TZEI 56 and TZEI 128 were moderately resistant to the pathogen with DS scores of 3.7, 4.2 and 3.8, respectively. However, TZEI 134, TZEI 268, TZEI 135 and TZEI 1 were susceptible to the pathogen. Furthermore, at 6WAI (10 weeks after planting), the same pattern was recorded for the inbred lines. For the DLA, TZEI 56, TZEI 60 and TZEI 86 had the least DS scores, whereas TZEI 120 and TZEI 135 had higher DS scores. At 10DAI, TZEI 86, TZEI 56 and TZEI 60 had the least severity score which indicated moderate resistance, while TZEI 124, TZEI 134, TZEI 135 and TZEI 1 had the highest severity score. Notably, similar reactions of the inbred lines were observed regardless of the method of evaluation either the DLA or the screenhouse evaluation.
DISCUSSION
Ten of each of the early and extra-early maturing inbreds were screened for resistance to E. turcicum and B. maydis independently through laboratory assay (DLA) and screenhouse screening to document the reaction of the inbred lines to the pathogens. The inoculation methods used for both screenhouse and DLA experiments facilitated the gradual spread of the pathogen with visible necrotic lesions characteristic of the pathogen from the point of inoculation to other parts of the plants. This facilitated the effective discrimination and identification of resistant genotypes based on the disease severity scales. The DLA, facilitated the rapid identification of EM and EEM maize inbreds with resistance to NCLB and SCLB, respectively. In addition to the identification of resistant genotypes, the screenhouse evaluations provided useful agronomic information on the genotypes which is a major deficiency of the DLA. The disease severity scores of the genotypes were found to be similar regardless of the method used. This underscores the role of the DLA as an effective rapid tool for the identification of disease-resistant genotypes to foliar diseases.
Northern and southern corn leaf blight caused by E. turcicum and B. maydis, respectively are major foliar diseases that affect maize production globally7,17. However, management through the application of fungicides is expensive and detrimental to the environment. Disease management through the use of resistant genotypes is effective, cost-efficient and adjudged as the most reliable option. Identification of NCLB and SCLB resistant genotypes is, therefore, crucial for the development of high-yielding hybrids for improved crop production and disease management7,30,38.
Several plant inoculation techniques including: Atomizing spore suspensions39, dropping a spore suspension into the whorl40, application of grinded symptomatic leaves41, combined spore suspension and inoculated sorghum grains either simultaneously or at five days interval42, have been reported. However, the inoculum production and whorl inoculation methods adopted from Badu-Apraku et al.7 and Bankole et al.15 were effective. Within five days of the sorghum grain inoculation, the pathogens (E. turcicum and B. maydis) sporulated and colonized the sterile sorghum. At the point of inoculation, the maize whorls were fully formed held the calibrated grains allotted to each plant and facilitated disease establishment. Additionally, the solid-grain inoculation technique allowed gradual progression of the disease through the crop’s developmental stages as symptoms evolved from the spot of inoculation to the new leaves. The gradual spread of the pathogen from the point of inoculation has been identified as an important component for monitoring disease spread as well as classifying the reaction of the plant as either susceptible, tolerant or resistant15.
In addition, the DLA, facilitated rapid identification EM and EEM maize inbreds with resistance to NCLB and SCLB, respectively. This was similar to the reports of Ogunsanya et al.36, where DLA was found to be a rapid and effective technique for investigating host-parasite interactions, identifying resistant genotypes, screening for broad-spectrum resistance to plant diseases in a variety of crops. Given that leaf age significantly affects pathogen infection43,44, the present study adjusted for the use of disease free 4-weeks-old leaves. Additionally, using DLA offered the benefit of managing environmental conditions, preventing biotic stress brought on by other microbes and could be a more accurate way to test for NCLB and SCLB resistance than the screenhouse technique.
The results of the screenhouse experiment were similar to those of the laboratory using DLA. Both methods identified TZEEI 8, TZEEI 20 and TZEEI 179, as the resistant extra-early maturing inbred lines to B. maydis while TZEEI 158 and TZEEI 32 were identified as susceptible. Similarly, TZEI 56, TZEI 60 and TZEI 86 with the least DS score, were identified as NCLB-resistant inbreds whereas TZEI 1, TZEI 120, TZEI 135 and TZEI 269 had the highest DS score and were susceptible to E. turcicum by both methods. These results validated the DLA as a rapid method for identification of resistant germplasm offering significant reduction in time, financial resources and labor requirements. Although, the DLA did not proffer information on the agronomic potentials of the inbred lines, it could be used as an initial screening method for studies involving large number of germplasm while the selected germplasm from the initial study can be subjected to screenhouse or field trials.
The present study also documented the agronomic potentials of the early and extra-early maize inbreds under artificial B. maydis and E. turcicum inoculations in screenhouse conditions. For example, under B. maydis inoculations, TZEEI 179 which was identified with SCLB-resistance had good agronomic traits with plant and ear aspect ratings of 4 as well as significantly low grain weight. The inbred is a good source for disease resistance which could be combined with other inbred with better agronomic properties for the development of hybrids with high productivity and resistance to diseases. Additionally, TZEEI 86 with be better phenotypic appeal and higher grain weight compared to TZEEI 179 was tolerant to the SCLB disease. Tolerance was observed among the extra-early maturing inbreds rather than resistance. Most inbreds with higher disease severity scores had higher grain weight (TZEEI 157 and TZEEI 158). This may be due to the minute lesion produced by symptomatic plants45, which may not have severe influence on the photosynthetic ability of the infected plants unlike NCLB that produces large and elongated lesion on affected plant parts. The reduced effect on grain yield may also be traced to the race of the pathogen as racial discrimination among the pathogen have known to produce different effects with varying severity on infected plants16.
Early maturing inbreds identified with NCLB-resistance by both screenhouse and DLA evaluations combined were also found to possess good agronomic traits and grain weight e.g., For example, TZEI 56, TZEI 60 and TZEI 86. Similarly, the inbred lines susceptible to NCLB (e.g., TZEI 120 and TZEI 1) were also found to have significantly lower grain weight. The NCLB causes significant loss of photosynthetic apparatus of infected plants40, with attendant grain yield losses . The screenhouse experiments allowed the identification of plant genotypes with disease resistance as well as desirable agronomic traits.
CONCLUSION
The TZEEI 179, TZEEI 8 and TZEEI 20 were identified as SCLB-resistant inbreds while TZEI 56, TZEI 60, TZEI 86 and TZEI 128 were found to be NCLB-resistant genotypes using both methods. The DLA is an efficient technique for identifying disease-resistant EM and EEM genotypes and could facilitate rapid development of disease-resistant maize hybrids and open-pollinated varieties. Adopting the disease-resistant genotypes would reduce crop losses, increase farmer’s income and ensure food security. Detached leaf assay should be adopted for rapid screening of genotypes in disease-resistance studies for maize foliar diseases prior to field evaluations. Additionally, the identified inbred lines are genetic resources that could facilitate the development of disease-resistant maize hybrids and population for increased maize production.
SIGNIFICANCE STATEMENT
Maize is susceptible to several biotic stresses and diseases that limit its maximum production and productivity. Host plant resistance has been adjudged the best option among the various options for disease management. This present study compared the effectiveness of a rapid laboratory assay; the detached leaf assay (DLA) with the traditional screenhouse screening method for rapid identification of disease-resistant maize genotypes. Both methods identified similar early and extra-early maturing maize genotypes with resistance to two devastating foliar diseases. The study demonstrated the effectiveness of DLA for rapid identification of disease-resistant maize genotypes. This would facilitate accelerated genetic gains through early selection for the development of foliar disease-resistant maize varieties. Adoption of these varieties would ensure food security and increase farmer’s income.
ACKNOWLEDGMENT
The authors appreciate Dr. Baffour Badu-Apraku and IITA Maize Improvement Program for releasing the seeds used for this study.
REFERENCES
- FAO, 2018. The Future of Food and Agriculture: Trends and Challenges. Food & Agriculture Org., Rome, Italy, ISBN: 9789251095515, Pages: 180.
- FAO, 2021. World Food and Agriculture-Statistical Yearbook 2021. FAO, Rome, Italy, ISBN: 978-92-5-134332-6, Pages: 368.
- Singh, C., 2002. Modern Techniques of Raising Field Crops. Oxford and IBH, New Delhi, ISBN: 9788120401358, Pages: 523.
- Magar, P.B., G.B. Khatri-Chhetri, S.M. Shrestha and T.R. Rijal, 2018. Screening of maize genotypes against Southern leaf blight (Bipolaris maydis) during summer in Rampur, Chitwan, Nepal. J. Inst. Agric. Anim. Sci., 33-34: 115-122.
- Kumari, R., A.K. Singh and V.K. Sharma, 2018. Genetic divergence studies for morpho-agronomical traits in maize (Zea mays L.) inbreds. Int. J. Curr. Microbiol. Appl. Sci., S7: 3589-3594.
- Oyekunle, M. and B. Badu-Apraku, 2014. Genetic analysis of grain yield and other traits of early-maturing maize inbreds under drought and well-watered conditions. J. Agron. Crop Sci., 200: 92-107.
- Badu-Apraku, B., F.A. Bankole, B.S. Ajayo, M.A.B. Fakorede and R.O. Akinwale et al., 2021. Identification of early and extra-early maturing tropical maize inbred lines resistant to Exserohilum turcicum in Sub-Saharan Africa. Crop Prot., 139.
- Schoch, C.L., S. Ciufo, M. Domrachev, C.L. Hotton and S. Kannan et al., 2020. NCBI taxonomy: A comprehensive update on curation, resources and tools. Database, 2020, 10.1093/database/baaa062
- Kumar, D. and A. Jhariya, 2013. Nutritional, medicinal and economical importance of corn: A mini review. Res. J. Pharm. Sci., 2: 7-8.
- Badu-Apraku, B. and M.A.B. Fakorede, 2017. Maize in Sub-Saharan Africa: Importance and Production Constraints. In: Advances in Genetic Enhancement of Early and Extra-Early Maize for Sub-Saharan Africa, Badu-Apraku, B. and M.A.B. Fakorede (Eds.), Springer, Cham, Switzerland, ISBN: 978-3-319-64852-1, pp: 3-10.
- Lans, C.A., 2006. Ethnomedicines used in Trinidad and Tobago for urinary problems and diabetes mellitus. J. Ethnobiol. Ethnomed., 2.
- Shah, T.R., K. Prasad and P. Kumar, 2016. Maize-A potential source of human nutrition and health: A review. Cogent Food Agric., 2.
- Kennett, D.J., K.M. Prufer, B.J. Culleton, R.J. George and M. Robinson et al., 2020. Early isotopic evidence for maize as a staple grain in the Americas. Sci. Adv., 6.
- García-Lara, S. and S.O. Serna-Saldivar, 2019. Corn History and Culture. In: Corn: Chemistry and Technology, Serna-Saldivar, S.O. (Ed.), Elsevier, Netherlands, ISBN: 978-0-12-811971-6, pp: 1-18.
- Bankole, F.A., B. Badu-Apraku, A.O. Salami, T.D.O. Falade, R. Bandyopadhyay and A. Ortega-Beltran, 2022. Identification of early and extra-early maturing tropical maize inbred lines with multiple disease resistance for enhanced maize production and productivity in Sub-Saharan Africa. Plant Dis., 106: 2638-2647.
- Bankole, F.A., B. Badu-Apraku, A.O. Salami, T.D.O. Falade, R. Bandyopadhyay and A. Ortega-Beltran, 2023. Variation in the morphology and effector profiles of Exserohilum turcicum isolates associated with the northern corn leaf blight of maize in Nigeria. BMC Plant Biol., 23.
- Munkvold, G.P. and D.G. White, 2016. Compendium of Corn Diseases. 4th Edn., American Phytopathological Society, USA, ISBN: 978-0-89054-494-5, Pages: 165.
- Gong, F., L. Yang, F. Tai, X. Hu and W. Wang, 2014. “Omics” of maize stress response for sustainable food production: Opportunities and challenges. OMICS: J. Integr. Biol., 18: 714-732.
- Jibril, S.M., B.H. Jakada, A.S. Kutama and H.Y. Umar, 2016. Plant and pathogens: Pathogen recognision, invasion and plant defense mechanism. Int. J. Curr. Microbiol. Appl. Sci., 5: 247-257.
- Knogge, W., 1996. Fungal infection of plants. Plant Cell, 8: 1711-1722.
- Nazarov, P.A., D.N. Baleev, M.I. Ivanova, L.M. Sokolova and M.V. Karakozova, 2020. Infectious plant diseases: Etiology, current status, problems and prospects in plant protection. Acta Naturae, 12: 46-59.
- Ali, F., Q. Pan, G. Chen, K.R. Zahid and J. Yan, 2013. Evidence of multiple disease resistance (MDR) and implication of meta-analysis in marker assisted selection. PLoS ONE, 8. https://doi.org/10.1371/journal.pone.0068150
- Manamgoda, D.S., L. Cai, E.H.C. McKenzie, P.W. Crous and H. Madrid et al., 2012. A phylogenetic and taxonomic re-evaluation of the Bipolaris-Cochliobolus-Curvularia complex. Fungal Diversity, 56: 131-144.
- Mubeen, S., M. Rafique, M.F.H. Munis and H.J. Chaudhary, 2017. Study of southern corn leaf blight (SCLB) on maize genotypes and its effect on yield. J. Saudi Soc. Agric. Sci., 16: 210-217.
- Akinwale, R.O. and A.O. Oyelakin, 2018. Field assessment of disease resistance status of some newly-developed early and extra-early maize varieties under humid rainforest conditions of Nigeria. J. Plant Breed. Crop Sci., 10: 69-79.
- Aregbesola, E., A. Ortega-Beltran, T. Falade, G. Jonathan, S. Hearne and R. Bandyopadhyay, 2020. A detached leaf assay to rapidly screen for resistance of maize to Bipolaris maydis, the causal agent of southern corn leaf blight. Eur. J. Plant Pathol., 156: 133-145.
- Manching, H.C., P.J. Balint-Kurti and A.E. Stapleton, 2014. Southern leaf blight disease severity is correlated with decreased maize leaf epiphytic bacterial species richness and the phyllosphere bacterial diversity decline is enhanced by nitrogen fertilization. Front. Plant Sci., 5.
- Horwitz, B.A., B.J. Condon and B.G. Turgeon, 2013. Cochliobolus heterostrophus: A Dothideomycete Pathogen of Maize. In: Genomics of Soil- and Plant-Associated Fungi, Horwitz, B.A., P.K. Mukherjee, M. Mukherjee and C.P. Kubicek (Eds.), Springer, Berlin, Heidelberg, Germany, ISBN: 978-3-642-39339-6, pp: 213-228.
- Smith, D.R. and R. Toth, 1982. Histopathological changes in resistant (rhm) corn inoculated with Helminthosporium maydis race O. Mycopathologia, 77: 83-88.
- Byrnes, K.J., 1989. Relationships between yield of three maize hybrids and severity of southern leaf blight caused by race O of Bipolaris maydis. Plant Dis., 73: 834-840.
- Sibiya, J., P. Tongoona, J. Derera and I. Makanda, 2013. Smallholder farmers' perceptions of maize diseases, pests, and other production constraints, their implications for maize breeding and evaluation of local maize cultivars in KwaZulu-Natal, South Africa. Afr. J. Agric. Res., 8: 1790-1798.
- Human, M.P., I. Barnes, M. Craven and B.G. Crampton, 2016. Lack of population structure and mixed reproduction modes in Exserohilum turcicum from South Africa. Phytopathology, 106: 1386-1392.
- Bernardo, R., 2014. Essentials of Plant Breeding. Stemma Press, USA, ISBN 13: 9780972072427.
- Wiesner-Hanks, T. and R. Nelson, 2016. Multiple disease resistance in plants. Annu. Rev. Phytopathol., 54: 229-252.
- Balint-Kurti, P.J. and G.S. Johal, 2009. Maize Disease Resistance. In: Handbook of Maize: Its Biology, Bennetzen, J.L. and S.C. Hake (Eds.), Springer, New York, ISBN: 978-0-387-79418-1, pp: 229-250.
- Ogunsanya, O.M., M.A. Adebisi, A.R. Popoola, C.G. Afolabi and O. Oyatomi et al., 2024. Morphological, pathological and phylogenetic analyses identify a diverse group of Colletotrichum spp. causing leaf, pod, and flower diseases on the orphan legume African yam bean. bioRxiv.
- Rife, T.W. and J.A. Poland, 2014. Field book: An open-source application for field data collection on android. Crop Sci., 54: 1624-1627.
- Kump, K.L., P.J. Bradbury, R.J. Wisser, E.S. Buckler and A.R. Belcher et al., 2011. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet., 43: 163-168.
- Ahangar, M.A., Z.A. Bhat, F.A. Sheikh, Z.A. Dar, A.A. Lone, K.S. Hooda and M. Reyaz, 2016. Pathogenic variability in Exserohilum turcicum and identification of resistant sources to turcicum leaf blight of maize (Zea mays L.). J. Appl. Nat. Sci., 8: 1523-1529.
- Weems, J.D. and C.A. Bradley, 2018. Exserohilum turcicum race population distribution in the North Central United States. Plant Dis., 102: 292-299.
- Debela, M., M. Dejene and W. Abera, 2017. Management of Turcicum leaf blight [Exserohilum turcicum (Pass.) Leonard Suggs] of maize (Zea mays L.) through integration of host resistance and fungicide at Bako, Western Ethiopia. Afr. J. Plant Sci., 11: 6-22.
- Wani, T.A., G.N. Bhat, M. Ahmad and A. Anwar, 2017. Integrated disease management of Turcicum leaf blight of maize caused by Exserohilum turcicum. Int. J. Curr. Microbiol. Appl. Sci., 6: 149-155.
- Degani, O. and G. Cernica, 2014. Diagnosis and control of Harpophora maydis, the cause of late wilt in maize. Adv. Microbiol., 4: 94-105.
- Weihmann, F., I. Eisermann, R. Becher, J.J. Krijger, K. Hübner, H.B. Deising and S.G.R. Wirsel, 2016. Correspondence between symptom development of Colletotrichum graminicola and fungal biomass, quantified by a newly developed qPCR assay, depends on the maize variety. BMC Microbiol., 16.
- Rossman, A.Y., D.S. Manamgoda and K.D. Hyde, 2013. (2234) Proposal to conserve the name Helminthosporium maydis Y. Nisik. & C. Miyake (Bipolaris maydis) against H. maydis Brond. and Ophiobolus heterostrophus (Ascomycota: Pleosporales: Pleosporaceae). TAXON, 62: 1332-1333.
How to Cite this paper?
APA-7 Style
Bankole,
F.A., Bamidele,
P.I., Adeoye,
F., Salami,
A.O. (2024). Rapid Screening Methods for Identification of Resistant Maize Inbreds to Fungal Foliar Pathogens. Asian Journal of Biological Sciences, 17(4), 741-753. https://doi.org/10.3923/ajbs.2024.741.753
ACS Style
Bankole,
F.A.; Bamidele,
P.I.; Adeoye,
F.; Salami,
A.O. Rapid Screening Methods for Identification of Resistant Maize Inbreds to Fungal Foliar Pathogens. Asian J. Biol. Sci 2024, 17, 741-753. https://doi.org/10.3923/ajbs.2024.741.753
AMA Style
Bankole
FA, Bamidele
PI, Adeoye
F, Salami
AO. Rapid Screening Methods for Identification of Resistant Maize Inbreds to Fungal Foliar Pathogens. Asian Journal of Biological Sciences. 2024; 17(4): 741-753. https://doi.org/10.3923/ajbs.2024.741.753
Chicago/Turabian Style
Bankole, Faith, Ayobami, Priscilla Iseoluwa Bamidele, Florence Adeoye, and Abiodun Olusola Salami.
2024. "Rapid Screening Methods for Identification of Resistant Maize Inbreds to Fungal Foliar Pathogens" Asian Journal of Biological Sciences 17, no. 4: 741-753. https://doi.org/10.3923/ajbs.2024.741.753

This work is licensed under a Creative Commons Attribution 4.0 International License.


