Growth Analysis and Characterization of Some Varieties of Capsicum Species in Akwa Ibom State, Nigeria
| Received 30 Jul, 2024 |
Accepted 19 Sep, 2024 |
Published 31 Dec, 2024 |
Background and Objective: Peppers are among the vegetables that provide a rich source of various bioactive compounds with potential health-improving properties. However, their occurrence and distribution differ by genotype and maturity phase in plants. Morphogenetic analysis of some varieties of Capsicum species was assessed. Materials and Methods: Mature Capsicum fruit varieties were collected from 13 areas across, the state. The seeds were removed, sun-dried for 3 days and stored for 2 weeks at room temperature (15-25̊C) in polythene bags before planting. Leaf and shoot lengths were measured at 7 days intervals till maturity. The mature fruits were harvested for genetic and capsaicin analysis. Means, standard errors and Two-way Analysis of Variance (ANOVA) were carried out using GraphPad Prism (6.0). However, a probability level of p = 0.05 was considered statistically significant. Cluster analysis (Dendrogram) to show the phylogeny of the Capsicum varieties was done using Minitab software (version 17). Results: The varietal phenotypic characters studied varied significantly (p<0.05). On day 70, red long dry pepper had the largest leaf length (16.030±0.44 cm) and shoot length (23.880±1.03 cm) while yellow pepper had the least leaf length (4.580±0.24 cm) and shoot length (7.870±0.24 cm). Evolutionary relationship using DNA revealed that all the varieties belonged to one clade except the red long dry pepper. Evolutionary relationship with capsaicin gene showed that red long plumpy and green big round peppers belonged to the same clade as Capsicum annum and Capsicum frutescens and yellow, red long slender, shombo and sub-shombo peppers belonged to the same clade with Capsicum frutescens while red long dry and round peppers belong to one clade. Conclusion: This study shows that Capsicum varieties exhibit genetic similarities and variations with implications in genetic hybridization, taxonomy and conservation.
| Copyright © 2024 Archibong et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Plants containing multiple bioactive compounds are becoming more and more popular due to their ability to enhance human health and nutrition1,2. Identifying these plant species with medicinal significance accurately is the initial and fundamental stage in any improvement initiative. This enables the dependable and efficient selection of appropriate parental genotypes (ensuring genuine purity and quality) in plant breeding schemes for diverse nutritional and pharmacological objectives3.
The Solanaceae family includes the genus Capsicum, which is known by various universal English names like hot pepper, chile pepper, chili, sweet pepper and bell pepper4. There are approximately 27 species of Capsicum, with five of them being extensively grown. These include C. annuum L., C. chinensis Jacq., C. frutescens L., C. pubescens R. and C. baccatum L.4. Capsicum spp. are cultivated in tropical and subtropical regions of the world and they are significant and well-liked vegetables and spices5. Not only are they highly valued for their economic significance, but they are also appreciated for their abundant nutritional benefits. Pepper fruits are packed with various bioactive phytochemicals such as flavonoids, carotenoids, phenolics and other antioxidants6. In addition to being used as food flavorings and for their nutritional value, the spicy Capsicum spp. play an important role in the field of medicine due to their capsaicin content and are currently utilized for various therapeutic reasons. The active substances in red peppers that contribute to their medicinal and pharmaceutical applications are the spicy alkaloids known as capsaicinoids7.
Despite the widespread popularity of pepper across all agroecological zones, the level of genetic variation within the species is crucial for its long-term cultivation and ability to adapt to diverse agroecologies. Greater diversity within the population is advantageous for breeders in creating high-quality cultivars by carefully selecting exceptional parent plants. Therefore, it is crucial to comprehend the genetic variability of a population using both morphological and molecular markers. This understanding is essential for developing effective strategies for germplasm conservation and breeding purposes8. Pepper genotypes have been distinguished and grouped based on phenotypic traits such as fruit weight, flower color, fruit shape and plant height9. Describing and categorizing germplasm using phenotypic traits is a crucial initial stage in any characterization program10. Nonetheless, research has indicated that assessing the morphology of peppers to identify genetic variations is susceptible to environmental influences and may struggle to differentiate closely related individuals11.
Thus, the study aimed to evaluate the morphogenetic properties of some varieties of Capsicum spp in Akwa Ibom State, Nigeria.
MATERIALS AND METHODS
Study area: Specimens for this study were collected from 13 Local Government Areas across the three Senatorial Districts (Uyo, Ikot Ekpene and Eket) in Akwa Ibom State. The villages and their respective coordinates where the species were collected are presented in Table 1. Akwa Ibom State is located in the coastal Southern part of Nigeria. It lies between Latitudes 4°32'N and 5°33'N and Longitudes 7°25'E and 8°25'E. The state is bordered on the east by Cross River State, on the west by Rivers State and Abia State, on the north by Ebonyi State and on the south by Bight of Biafra of the Atlantic Ocean. It has a total land area of about 7,081 km2. The area has characteristically two seasons, dry and wet seasons. The dry season of the area occurs between November and March, while the wet season stretches between April and October. Rainfall is heavy and ranges from 3,000 mm along the coast to 2,000 mm on the north fringe. The mean temperature of the area is usually uniformly high throughout the year with slight variation between 25 and 28°C. Relative humidity is usually high, ranging between 75 and 85%. The study duration is February-July, 2024.
Collection and propagation of plant materials: Mature fruits of various Capsicum varieties (nine) were obtained from 13 Local Government Areas across the three Senatorial Districts (Uyo, Ikot Ekpene and Eket) in Akwa Ibom State. For Uyo Senatorial District, samples were collected from Urua Akpan Andem (Uyo), Urua Afaha Offiong (Nsit Ibom), Urua Odot (Nsit Atai) and Urua T-junction Oboetim (Itu). For Ikot Ekpene Senatorial District, samples were collected from Urua Attor (Ikot Ekpene), Urua Nkari (Ini), Urua Udonkok

|

|

|
| Table 1: | Coordinates of the study areas | |||
| Local governments | Latitude (°N) | Longitude (°E) | Altitude (ft) |
| Uyo Senatorial District | |||
| Uyo | 5.015251 | 7.925669 | 278 |
| Nsit Ibom | 4.869738 | 7.903226 | 205 |
| Itu | 5.200302 | 7.977335 | 86 |
| Ikot Ekpene Senatorial District | |||
| Ikot Ekpene | 5.18432 | 7.716421 | 355 |
| Ini | 5.387142 | 7.67653 | 451 |
| Etim Ekpo | 4.964826 | 7.593536 | 264 |
| Ukanafun | 4.87692 | 7.565049 | 164 |
| Abak | 4.981349 | 7.781417 | 219 |
| Eket Senatorial District | |||
| Mbo | 4.655009 | 8.253211 | 130 |
| Eket | 4.657084 | 7.958585 | 137 |
| Ibeno | 4.544361 | 7.990957 | 59 |
| Eastern Obolo | 4.522695 | 7.754939 | 66 |
| Mkpat Enin | 4.65942 | 7.781625 | 138 |
(Ukanafun), Urua Obo (Etim Ekpo) and Urua Abak (Abak). For Eket Senatorial District, samples were obtained from Badans Farms (Eket), Urua Enwang (Mbo), Okoroiti (Eastern Obolo), Urua Ukam (Mkpat Enin) and Urua Iwuokpom (Ibeno) (Fig. 1-9). Seeds in the fruits were first removed, sun-dried (for 3 days) and stored (for 2 weeks) at room temperature of about 15-25°C in polythen bags before planting. Planting was done using labeled plastic pots filled with soil (3 kg) in the Green House of the Department of Botany and Ecological Studies, University of Uyo, between April 2020 and September 2020. Mature fruits of the Capsicum varieties were harvested and analyzed in the laboratory qualitatively and quantitatively for phytochemicals. Antioxidants and capsaicin were also analyzed for the Capsicum fruits.

|

|

|

|

|

|
Measurements of phenotypic characters of Capsicum varieties
Shoot length: A meter rule (cm) was used in measuring the shoot of the plant from the ground level to the apex. These measurements were taken at intervals of seven (7) days.
Leaf length: A meter rule (cm) was used in measuring the leaf from the node to the apex. These measurements were taken at the intervals of seven (7) days.
DNA extraction: The DNA extraction was done using Genomic DNA Kit (Plant) by Geneaid. The process for extracting the DNA from the Capsicum leaves involved the following steps; tissue dissociation, lysis, DNA binding and DNA Elution.
Tissue dissociation: The 100 mg of fresh or young Capsicum leaf was cut off. The sample was ground in liquid nitrogen in a mortar with a pestle to a fine powder and transferred to 1.5 mL microcentrifuge tube. Four hundred microlitre (400 μL) of GP1 buffer and 5 μL of RNase A were added to the sample tube and mixed by vortex (Fisher Scientific). The sample was incubated at 60°C for 10 min. During incubation, the tube was inverted at every 5 min.
Lysis: The 100 μL of GP2 buffer was added to the sample lysate, mixed by vortex and incubated on ice for 3 min.
Filtration: A Filter Column was placed in a 2 mL collection tube and the mixture was transferred to the Filter Column. The mixture was centrifuged for 1 min at 1,000 × g and the Filter Column was discarded. The required Elution Buffer (200 μL per sample) was preheated to 60°C for later use during DNA elution.
DNA binding: The supernatant was carefully transferred from the 2 mL collection tube to a new 1.5 mL microcentrifuge tube. Exactly 1.5 volume of isopropanol was added and vortexed immediately for 5 sec. Column was placed in a 2 mL collection tube. Seven hundred microlitre (700 μL) of the mixture (and any remaining precipitate) was transferred to the GD Column. It was centrifuged at 14-16,000 × g for 2 min.
Washing: The 400 μL of W1 buffer was added to the GD column and centrifuged at 14-16,000 × g for 30 sec. The flow-through was discarded and the GD column was placed back in the 2 mL collection tube. Six hundred microlitre (600 μL) of ethanol was added to the GD column and centrifuged at 14-16,000 × g for 30 sec. The flow-through was discarded and the GD column was placed back in the 2 mL collection tube. It was centrifuged further for 3 min at 14-16,000 × g to dry the column matrix.
DNA elution: The dried GD column was transferred to a clean 1.5 mL microcentrifuge tube. One hundred microlitre (100 μL) of pre-heated Elution Buffer was added to the center of the column matrix and allowed to stand for 3-5 min to ensure the Elution Buffer was completely absorbed. It was centrifuged at 14-16, 000 × g for 30 sec to elute the purified DNA.
| Table 2: | Primer sequences used in the study | |||
| Primer name | Sequence (5’-3’) | Amplicon size (bp) |
| CAP F | CCGGTGAAATACCCGGAGAG | 840 |
| CAP R | TGAGACCATTTCCTCCCAAG | |
| trnH-psbA F | GTTATGCATGAACGTAATGCTC | 558 |
| trnH-psbA R | CGCGCATGGTGGATTCACAATCC |
DNA quantification: The DNA concentration was determined using a Spectrophotometer (Gene Quant Pro). The absorbance of total genomic DNA (gDNA) was quantified by measuring optical density (OD) at 260 and 280 nm.
Gel electrophoresis: The presence and quality of gDNA was also evaluated by agarose gel electrophoresis. The DNA was accessed on 1.5% agarose gel. Electrophoresis was conducted in a 1X TAE (Tris-base glacial acetic acid, EDTA) buffer at 120 volts for 20 min. The gel was stained with 7 μL of SafeView dye. The gel was visualized under an EV transilluminator (Accuris SmartDoc 2.0).
PCR amplification of the genes: The PCR was conducted separately for each primer, that is CAP primers and trnh-psbA genes (Table 2). The PCR master mix contained PCR amplification buffer, MgCl, DMSO, DNTPs and Taq polymerase. Other reagents include CAP primers (forward and reverse), ultra-pure molecular biology grade water H2O and template DNA.
Cocktail mix: The PCR final reaction volume was made up to 20 μL, using 4 μL Solisbiodyne master mix, 0.5 μL forward and reverse primer, 2 μL of sample DNA and 13 μL nuclease-free water.
PCR conditions: The PCR reaction conditions conducted at BIO-RAD thermocycler, were the following: Initial denaturation at 95°C for 30 sec, 30 cycles of denaturation at 95°C for 30 sec, annealing at 51°C for 30 sec, initial elongation at 68°C for 1 min and final extension at 75°C for 5 min. Amplicons were separated on 1.5% agarose gel electrophoresis for 20 min at 120 V. A DNA ladder of 100 bp was used as the molecular weight standard.
Sequencing and sequence analysis: The PCR products obtained with TRNH and CAP primers were purified and sequenced in forward and reverse directions to determine the sequence of nucleotides for both TRNH and Capsaicin genes in the 9 pepper varieties. Sanger sequencing was carried out on ABI Prism 3130X1 Genetic Analyzer (Applied Biosystems) and BigDye terminator V3.1 kit (Applied Biosystems Inc.). The sequencing reaction was set up in 10.0 μL volume using 1.0 μL of 1 mL BigDye terminator√3.1 (Applied Biosystems), 1.5 μL of BigDye buffer (5×), 1.5 μL of 2.5 pmole of the reverse or the forward primer, 1.0 μL of the purified template DNA, generated from the sequencing reaction products and 5.0 μL of GIBCO water (Invitrogen Corporation). The sequencing PCR profile has an initial denaturation of 96°C for 1 min after performing a rapid ramp to 96°C and was subjected to 25 cycles of the following program: 96°C for 10 sec, 50°C for 5 sec, 60°C for 4 min and performed rapid thermal ramp for 4°C and held forever. The sequencing reaction products were cleaned up through an ethanol purification method consisting of 1.0 μL 125 mM EDTA, 1.0 μL 3M NaOAc2 and 25.0 μL 100% ethanol, before the addition of 9 μL of Hi-Di formamide (Applied Biosystems) to each of the purified sequencing PCR product. The mixtures were denatured for 5 min at 95°C prior to the sample analysis on an ABI Prism 3130X1 Genetic automated sequencer (Applied Biosystems) using the default settings. Obtained raw sequences were edited by trimming both ends and removing bad chromatograms. Nucleotide sequences were aligned by ClustalW. The BLASTn algorithm was conducted on NCBI with aligned sequences for each variety to determine its sequence similarity and identity.
Evolution relationship of the Capsicum varieties: The UPGMA method was used to infer the evolutionary history12. The optimal tree has been displayed. Next to the branches, the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are also shown13. The Tamura and Nei14 method was used to calculate the evolutionary distances, which are expressed as the number of base substitutions per site. A total of 9 nucleotide sequences were included in this analysis. The codon positions considered were 1st+2nd+3rd+Noncoding. Any ambiguous positions were deleted for each sequence pair using the pairwise deletion option. The final dataset comprised 537 positions. The evolutionary analyses were performed using MEGA X15.
Statistical data analyses: Means, standard errors and Two-way Analysis of Variance (ANOVA) were carried out using GraphPad Prism (6.0). However, a probability level of p = 0.05 was considered statistically significant. Cluster analysis (Dendrogram) to show the phylogeny of the Capsicum varieties was done using Minitab software (version 17).
RESULTS
Phenotypic characters of the Capsicum varieties
Leaf length: Data on leaf lengths of the Capsicum varieties are presented in Table 3. Seven days after planting, there were no leaves for measurement. On the 14 day, Sub-Shombo pepper and yellow pepper had the highest and least leaf length values of 2.970±0.22 and 1.400±0.09 cm, respectively. Red long plumpy pepper recorded the highest leaf length values on days 21 (3.230±0.24 cm), 28 (4.480±0.30 cm) and 35 (4.890±0.42 cm) while yellow pepper had the least leaf length values on days 21 (1.580±0.05 cm), 28 (1.820±0.08 cm) and 35 (1.960±0.19 cm). On days 42, 49 and 56, round pepper had the highest leaf length values of 7.070±0.68, 8.670±0.62 and 10.250±0.61 cm while yellow pepper recorded the least values of 2.470±0.19, 3.030±0.20 and 3.520±0.20 cm on the aforementioned days. On days 63 and 70, red long dry pepper recorded the highest leaf length values of 12.730±0.55 and 16.030±0.44 cm while the least leaf length values of 3.890±0.19 and 4.580±0.24 cm were observed in yellow pepper. The values obtained for the leaf lengths in the varieties for the respective days differed significantly (p<0.05) (Table 3).
Shoot length: The data on shoot lengths of the Capsicum varieties are presented in Table 4. No data were collected on the seventh day after planting. Red long plumpy pepper had the highest values for shoot length on day 14 (3.890±0.15 cm) while green small round pepper had the least value of 2.720±0.29 cm. Red long dry pepper had the highest shoot length values of 5.380±0.25, 6.100±0.27 and 8.220±1.18 cm on days 21, 28 and 35, while green small round pepper had the least shoot length values of 3.050±0.32, 3.210±0.27 and 4.770±0.27 cm for days 21, 28 and 35, respectively. Red long plumpy pepper also had highest values for shoot length on days 42 (12.120±1.08 cm), 49 (14.000±1.06 cm) and 56 (15.640±1.31 cm) while the least values for shoot length on days 42 (5.660±0.26 cm), 49 (6.260±0.26 cm) and 56 (6.810±0.24 cm) were observed in yellow pepper. Red long dry pepper, on days 63 and 70, had the highest shoot length values of 18.760±1.13 and 23.880±1.03 cm while yellow pepper had the least values of 7.320±0.24 (day 63) and 7.870±0.24 cm (day 70). However, the values obtained for the shoot lengths in the varieties across the respective days differed significantly (p<0.05) (Table 4).
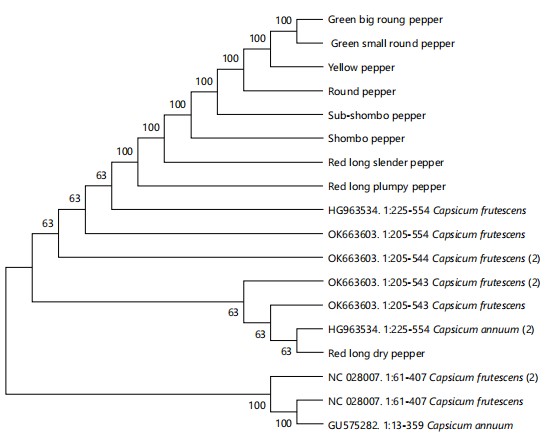
Evolutionary relationship of the Capsicum varieties studied: The evolutionary relationship of the Capsicum varieties studied is shown in Fig. 10. Phylogenetic tree showed that the Capsicum varieties used in the study clustered in one clade except for the red long dry pepper, which was in a different clade. Other sequences that were in the first clade include Capsicum frutescens and Capsicum annum. From the BLASTn analysis (Table 5), green small round pepper, green big round pepper and round pepper had a 99% similarity with Capsicum annum. Yellow, shombo, red long slender and red long plumpy peppers had 99% similarity with Capsicum frutescens; sub-shombo pepper had a 97% similarity with Capsicum frutescens while red long dry pepper had a 100% similarity with Capsicum frutescens.
| Table 3: | Leaf lengths (cm) of the Capsicum varieties studied |
|||
| Varieties | ||||||||||
| Days | Green small round pepper |
Green big round pepper |
Yellow pepper |
Round pepper |
Sub-Shombo pepper |
Shombo pepper |
Red long slender pepper |
Red long dry pepper |
Red long dry pepper |
|
| 7 | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | |
| 14 | 1.420±0.13a | 1.510±0.17a | 1.400±0.09a | 1.50±0.12a | 2.970±0.22b | 2.120±0.12b | 2.130±0.25b | 2.200±0.19b | 2.280±0.10b | |
| 21 | 1.590±0.19a | 2.180±0.14b | 1.580±0.05a | 2.070±0.04b | 2.160±0.06b | 3.320±0.07c | 2.710±0.23b | 3.230±0.24c | 2.420±0.16b | |
| 28 | 1.920±0.20a | 2.810±0.19a | 1.820±0.08a | 2.490±0.14a | 2.850±0.25a | 3.420±0.12b | 3.140±0.20b | 4.480±0.30c | 2.960±0.13a | |
| 35 | 3.030±0.21a | 3.120±0.26a | 1.960±0.19b | 3.890±0.50a | 4.010±0.29a | 3.110±0.26a | 3.160±0.20a | 4.890±0.42a | 3.540±0.38a | |
| 42 | 4.130±0.21a | 5.490±0.24b | 2.470±0.19c | 7.070±0.68d | 5.070±0.29b | 4.970±0.27b | 5.610±0.66b | 6.930±0.55d | 6.350±0.69d | |
| 49 | 5.270±0.21a | 6.200±0.26a | 3.030±0.20b | 8.670±0.62c | 5.690±0.38a | 5.930±0.51a | 6.430±0.60a | 7.530±0.59c | 6.900±0.72a | |
| 56 | 6.370±0.21a | 6.570±0.39a | 3.520±0.20b | 10.250±0.61c | 8.130±0.46d | 7.090±0.53d | 8.200±0.54d | 8.300±0.89d | 8.210±0.76d | |
| 63 | 7.450±0.30a | 7.050±0.40a | 3.890±0.19b | 10.800±0.59c | 9.110±0.46d | 8.110±0.38d | 9.430±0.34d | 10.330±0.95c | 12.730±0.55e | |
| 70 | 8.580±0.29a | 7.550±0.40a | 4.580±0.24b | 11.810±0.51c | 9.020±0.99a | 8.540±0.40a | 10.380±0.35d | 12.350±0.96c | 16.030±0.44e | |
| Means with different superscripts along the same row are significantly different (p<0.05) | ||||||||||
| Table 4: | Shoot lengths (cm) of the Capsicum varieties studied | |||
| Varieties | ||||||||||
| Days | Green small round pepper |
Green big round papper |
Yellow pepper |
Round pepper |
Sub-Shombo pepper |
Shombo pepper |
Red long slender pepper |
Red long plumpy pepper |
Red long dry pepper |
|
| 7 | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | 0.000±0.00a | |
| 14 | 2.720±0.29a | 3.460±0.19a | 3.810±0.16a | 3.630±0.19a | 3.260±0.27a | 3.860±0.50a | 3.850±0.41a | 3.890±0.15a | 3.750±0.31a | |
| 21 | 3.050±0.32a | 4.070±0.27b | 3.400±0.16a | 3.600±0.15a | 3.560±0.25a | 4.900±0.33b | 5.060±0.31b | 4.750±0.11b | 5.380±0.25b | |
| 28 | 3.210±0.27a | 4.350±0.22a | 4.390±0.21a | 4.460±0.15a | 3.900±0.29a | 5.470±0.32b | 5.350±0.27b | 5.430±0.26b | 6.100±0.27b | |
| 35 | 4.770±0.27a | 4.490±0.60a | 5.180±0.26a | 6.400±0.85b | 6.590±0.39b | 5.370±0.48a | 5.200±0.42a | 7.120±0.71b | 8.220±1.18b | |
| 42 | 6.370±0.24a | 6.370±0.60a | 5.660±0.26a | 9.450±0.97b | 7.270±0.55a | 8.320±0.55b | 8.010±0.60b | 12.120±1.08c | 11.01±1.13c | |
| 49 | 7.800±0.23a | 7.450±0.55a | 6.260±0.26a | 11.870±0.57b | 8.480±0.49a | 9.800±0.62c | 9.840±0.52c | 14.000±1.06d | 12.000±1.30b | |
| 56 | 9.300±0.23a | 7.980±0.65b | 6.810±0.24b | 15.610±0.73c | 10.440±0.44a | 11.290±0.49d | 11.970±0.40d | 15.640±1.31c | 14.990±1.54c | |
| 63 | 10.830±0.23a | 9.290±0.73a | 7.320±0.24b | 16.700±0.72c | 12.210±0.31d | 12.150±0.35d | 12.630±0.39d | 17.920±1.37e | 18.760±1.13e | |
| 70 | 12.330±0.23a | 10.750±0.72b | 7.870±0.24c | 18.200±0.62d | 13.550±0.25a | 12.760±0.31a | 13.500±0.37a | 21.310±1.11e | 23.880±1.03 | |
| Means with different superscripts along the same row are significantly different (p<0.05) | ||||||||||
|
| Table 5: | BLASTN analysis of aligned sequences of TRNH gene | |||
| Variety | Accession | Name | Percent identification | Alignment length |
| Green small round pepper | HG963534.1 | Capsicum annum | 99.7 | 557 |
| Green big round pepper | HG963534.1 | Capsicum annum | 99.39 | 554 |
| Yellow pepper | OK663603.1 | Capsicum frutescens | 99.12 | 570 |
| Round pepper | GU575282.1 | Capsicum annum | 98.56 | 534 |
| Sub-shombo pepper | OK663603.1 | Capsicum frutescens | 97.11 | 551 |
| Shombo pepper | NC028007.1 | Capsicum frutescens | 98.85 | 548 |
| Red long slender pepper | NC028007.1 | Capsicum frutescens | 99.42 | 585 |
| Red long plumpy pepper | OK663603.1 | Capsicum frutescens | 98.82 | 521 |
| Red long dry pepper | OK663603.1 | Capsicum frutescens | 100 | 576 |
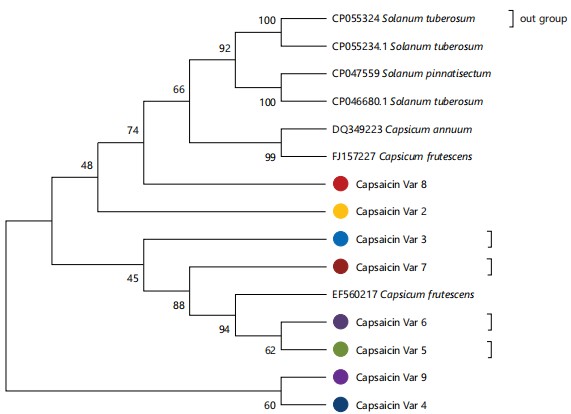
Evolutionary analysis of Capsicum varieties using Capsaicin gene by maximum likelihood method: A phylogenetic tree was also constructed using a maximum likelihood method with the Capsaicin gene sequences and data obtained from the genbank (Fig. 11). The Capsicum varieties were resolved to 3 phyletic clades based on their nucleotide diversity in the Capsaicin gene. Red long plumpy pepper and green big round pepper belonged to the same clade having a support value of 48% with other species such as Capsicum annum and Capsicum frutescens. Yellow pepper, red long slender pepper, shombo pepper and sub-shombo pepper formed a clade with another variety of Capsicum frutescens with a support value of 45% while red long dry pepper and round pepper were placed in the last clade with a 60% support value.
DISCUSSION
This study revealed significant variations (p<0.05) in the assessed morphological parameters (leaf length and shoot length) of the Capsicum varieties at different growing days. Similar findings have been reported by Adetula and Olakojo16, Thul et al.17 and Sharma et al.18. The leaf and shoot lengths of the Capsicum varieties varied directly with the growing days implying that, these parameters increased with increasing growing days. On the last day of growth (day 70), red long dry pepper and yellow pepper had the highest and least values for leaf and shoot lengths, respectively. Generally, the morphological parameters of these Capsicum varieties at day 70, followed this decreasing order; for leaf length (red long dry pepper>red long plumpy pepper>round pepper,>red long slender pepper>sub-shombo pepper>green small round pepper>shombo pepper>green big round pepper>yellow pepper) and shoot length (red long dry pepper>red long plumpy pepper>round pepper>sub-shombo pepper>red long slender pepper>shombo pepper>green small round pepper>green big round pepper>yellow pepper), respectively. These variations may be allied to differences in environmental factors, cultivars and genetic makeup of the varieties. This corroborated with the reports of Geleta et al.11 and Bermawie et al.19.
|
The amount of capsaicin also varied among the pepper varieties. A similar finding was reported by Canton-Flick et al.20 who asserted that capsaicin varies between species, among varieties within species, among plants within varieties, among fruits of the same plant and also among different parts of the same fruit. The variation in capsaicin levels in these varieties accounts for differences in their levels of spiciness, hotness, or pungency. This aligned with the report of Rahman and Inden21 that capsaicin is the compound responsible for the pungency of pepper fruits and their products. Campos et al.22 also maintained that the pungency and flavor are fruit attributes of Capsicum because capsaicin and dihydrocapsaicin are alkaloids that are responsible for 90 % of the intense organoleptic sensation of heat. According to Sathyanarayana23, these compounds are known for their therapeutic effects on gastric ulcers and rheumatoid arthritis. In this study, shombo pepper and yellow pepper had the highest and least capsaicin contents. The high capsaicin level in some varieties may be a pointer to their varieties with low capsaicin levels. The result of this study deviates from the findings of Zaki et al.24, who reported a high capsaicin level in green pepper over the red pepper. Nonetheless, the variations in capsaicin level of these varieties may be ascribed to factors, such as cultivar, genotype and stage of fruit development4.
Tewksbury et al.25 also added that environmental conditions such as intensity of light, temperature,nutrient requirements and water stress can potentially affect the accumulation of these alkaloids in this plant. In substantiating this, Sung et al.26 and Rahman and Inden21, reported that the highest capsaicin content in fruits was found in cultivars grown at a higher temperature and in nutrient-rich soils. The phenological stage of the fruit has also been reported to cause a variation in capsaicin levels of Capsicum27.
The grouping of the pepper varieties into the same cluster group is an indication of their varietal or genotypic relatedness. It further portrays similarities in their phylogeny. On the other hand, the grouping of pepper varieties into different cluster groups portrays their varietal or genotypic unrelatedness and distance. Sub-shombo pepper, which had a sub-cluster under it consisting of green small round and round peppers, was 99.43 % related to them (green small round and round pepper). Shombo and red long slender peppers formed a sub-cluster under green big round pepper with 99.47% relatedness to green big round pepper. Red long plumpy and red long dry peppers were related to each other at 99.35 %. These portray that these varieties, though slightly different in percentage relatedness, share a common progenitor. The placement of yellow pepper in a single group separated from every other variety portrays its phylogenetic distance. It may also indicate that yellow pepper originates from a stock different from the other varieties.
The clustering of green small round, green big round, yellow, round, sub-shombo, shombo, red long slender and red long plumpy peppers in one clade while assessing their evolutionary relationship using DNA may entail that these peppers originated from a common ancestral stock or had close genetic parents. It may also mean that these Capsicum varieties are closely related genetically. On the other hand, the segregation of red long dry pepper into a different clade may suggest that this variety of pepper is an out layer whose ancestral stock is different from the other varieties. Its segregation into a clade different from the other varieties may entail that red long dry pepper is genetically dissimilar from the other studied varieties. Based on the capsaicin gene analysis, a red long plumpy pepper and a green round pepper belonged to one clade alongside Capsicum annum and Capsicum frutescens in the genbank with a support value of 48 %. Yellow pepper, red long slender pepper, shombo pepper and sub-shombo pepper were segregated into one clade with Capsicum frutescens having a support value of 45 %. Be that as it may, It is pertinent to point out that these support values (48 and 45 %) are somewhat low and may depict a fairly genetically relatedness of the studied varieties with the genbank species based on the capsaicin gene. Conversely, the placement of the red long dry pepper and ground pepper in the same clade with a 60 % support value may portray a strong genetic resemblance with each other in terms of the capsaicin levels.
CONCLUSION
The evaluation of the genetic variation, which aids in proper delineation of plant species, is fundamental in agriculture as it helps to effectively conserve, manage and develop improved cultivars of plants endowed with bioactive compounds for various pharmacological uses. In this study, evaluation of genetic tools were used to characterize and classify the varieties and the species of Capsicum. Conclusively, this study shows that these Capsicum varieties exhibit a considerable amount of genetic similarities and variations based on their DNA.
SIGNIFICANCE STATEMENT
Peppers are among the vegetables that provide a rich source of various bioactive compounds with potential health-improving properties. However, their taxonomic and phylogenetic identities have not been clarified because of the overlap in morphological traits and bioactive compounds. Hence, there is a need to characterize the morphological parameters in different pepper varieties to establish their taxonomic identities and phylogenetic relatedness and improve the knowledge of their species relationship. This study is designed to assess the Morphogenetic analysis of some varieties of Capsicum species in Akwa Ibom State, Nigeria. Results showed phenotypic characters studied varied significantly amongst different varieties. Conclusively, these Capsicum varieties exhibit a considerable amount of genetic similarities and variations based on their DNA.
REFERENCES
- Idris, O.A., O.A. Wintola and A.J. Afolayan, 2017. Phytochemical and antioxidant activities of Rumex crispus L. in treatment of gastrointestinal helminths in Eastern Cape Province, South Africa. Asian Pac. J. Trop. Biomed., 7: 1071-1078.
- Unuofin, J.O., G.A. Otunola and A.J. Afolayan, 2017. Phytochemical screening and in vitro evaluation of antioxidant and antimicrobial activities of Kedrostis africana (L.) Cogn. Asian Pac. J. Trop. Biomed., 7: 901-908.
- Ganie, S.H., P. Upadhyay, S. Das and M.P. Sharma, 2015. Authentication of medicinal plants by DNA markers. Plant Gene, 4: 83-99.
- Bosland, P.W. and E. Votava, 2000. Peppers: Vegetable and Spice Capsicums. CABI, Wallingford, England, ISBN-13: 9780851993355, Pages: 204.
- Dimitrios, B., 2006. Sources of natural phenolic antioxidants. Trends Food Sci. Technol., 17: 505-512.
- Alvarez-Parrilla, E., L.A. de la Rosa, R. Amarowicz and F. Shahidi, 2011. Antioxidant activity of fresh and processed jalapeno and serrano peppers. J. Agric. Food Chem., 59: 163-173.
- Khan, F.A., T. Mahmood, M. Ali, A. Saeed and A. Maalik, 2014. Pharmacological importance of an ethnobotanical plant: Capsicum annuum (L). Nat. Prod. Chem., 28: 1267-1274.
- Lee, M.C., S.J. Oh, J.Y. Song, J. Lee and G.A. Lee et al., 2012. Evaluation of genetic diversity of red pepper landraces (Capsicum annuum L.) from Bulgaria using SSR markers. J. Korean Soc. Int. Agric., 24: 547-556.
- Weerakoon, S.R. and S. Somaratne, 2010. Agro-morphological characterization and relationships amoung mustard germplasm (Brassica juncea [L.] Czern & Coss) in Sri Lanka: A classification tree approach. J. Agric. Sci., 5: 89-97.
- Smith, J.S.C. and O.S. Smith, 1989. The description and assessment of distances between inbred lines of maize: II. The utility of morphological, biochemical, and genetic descriptors and a scheme for testing of distinctiveness between inbred lines. Maydica, 34: 151-161.
- Geleta, L., M.T. Labuschagne and C.D. Viljoen, 2005. Genetic variability in pepper (Capsicum annuum L.) estimated by morphological data and amplified fragment length polymorphism markers. Biodiversity Conserv., 14: 2361-2375.
- Sneath, P.H.A. and R.R. Sokal, 1973. Numerical Taxonomy: The Principles and Practice of Numerical Classification. W. H. Freeman, New York, ISBN: 9780716706977, Pages: 573.
- Felsenstein, J., 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution, 39: 783-791.
- Tamura, K. and M. Nei, 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. E, 10: 512-526.
- Kumar, S., G. Stecher, M. Li, C. Knyaz and K. Tamura, 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. E, 35: 1547-1549.
- Adetula, A.O. and S.A. Olakojo, 2006. Genetic characterization and evaluation of some pepper accessions Capsicum frutescens (L.): The Nigerian ‘Shombo’ collections. Am. Eurasian J. Agric. Environ. Sci., 1: 273-281.
- Thul, S.T., R.K. Lal, A.K. Shasany, M.P. Darokar and A.K. Gupta et al., 2009. Estimation of phenotypic divergence in a collection of Capsicum species for yield-related traits. Euphytica, 168: 189-196.
- Sharma, V.K., C.S. Semewal and S.P. Uniyal, 2010. Genetic variability and character association analysis in bell pepper (Capsicum annuum L.). J. Hortic. For., 2: 58-65.
- Bermawie, N., S. Wahyuni, R. Heryanto and I. Darwati, 2019. Morphological characteristics, yield and quality of black pepper Ciinten variety in three agro ecological conditions. IOP Conf. Ser.: Earth Environ. Sci., 292.
- Canto-Flick, A., E. Balam-Uc, J.J. Bello-Bello, C. Lecona-Guzmán and D. Solís-Marroquín et al., 2008. Capsaicinoids content in habanero pepper (Capsicum chinense Jacq.): Hottest known cultivars. Hortscience, 43: 1344-1349.
- Rahman, M.J. and H. Inden, 2012. Effect of nutrient solution and temperature on capsaicin content and yield contributing characteristics in six sweet pepper (Capsicum annuum L.) cultivars. J. Food Agric. Environ., 10: 524-529.
- Campos, M.R.S., K.R. Gomez, Y.M. Ordonez and D.B. Ancona, 2013. Polyphenols, ascorbic acid and carotenoids contents and antioxidant properties of habanero pepper (Capsicum chinense) fruit. Food Nutr. Sci., 4: 47-54.
- Satyanarayana, M.N., 2006. Capsaicin and gastric ulcers. Crit. Rev. Food Sci. Nutr., 46: 275-328.
- Zaki, N., A. Hasib, K.C. Eddine, F. Dehbi, H. El Batal and A. Ouatmane, 2017. Comparative evaluation of the phytochemical constituents and the antioxidant activities of five Moroccan pepper varieties (Capsicum annuum L.). J. Chem. Biol. Phys. Sci., 7: 1294-1306.
- Tewksbury, J.J., C. Manchego, D.C. Haak and D.J. Levey, 2006. Where did the chili get its spice? Biogeography of capsaicinoid production in ancestral wild chili species. J. Chem. Ecol., 32: 547-564.
- Sung, Y., Y.Y. Chang and N.L. Ting, 2005. Capsaicin biosynthesis in water-stressed hot pepper fruits. Bot. Bull. Acad. Sin., 46: 35-42.
- Materska, M., S. Piacente, A. Stochmal, C. Pizza, W. Oleszek and I. Perucka, 2003. Isolation and structure elucidation of flavonoid and phenolic acid glycosides from pericarp of hot pepper fruit Capsicum annuum L. Phytochemistry, 63: 893-898.
How to Cite this paper?
APA-7 Style
Archibong,
B.F., Akpabio,
K., Okon,
O.G. (2024). Growth Analysis and Characterization of Some Varieties of Capsicum Species in Akwa Ibom State, Nigeria. Asian Journal of Biological Sciences, 17(4), 796-808. https://doi.org/10.3923/ajbs.2024.796.808
ACS Style
Archibong,
B.F.; Akpabio,
K.; Okon,
O.G. Growth Analysis and Characterization of Some Varieties of Capsicum Species in Akwa Ibom State, Nigeria. Asian J. Biol. Sci 2024, 17, 796-808. https://doi.org/10.3923/ajbs.2024.796.808
AMA Style
Archibong
BF, Akpabio
K, Okon
OG. Growth Analysis and Characterization of Some Varieties of Capsicum Species in Akwa Ibom State, Nigeria. Asian Journal of Biological Sciences. 2024; 17(4): 796-808. https://doi.org/10.3923/ajbs.2024.796.808
Chicago/Turabian Style
Archibong, Bright, Frank, Kingsley Akpabio, and Okon Godwin Okon.
2024. "Growth Analysis and Characterization of Some Varieties of Capsicum Species in Akwa Ibom State, Nigeria" Asian Journal of Biological Sciences 17, no. 4: 796-808. https://doi.org/10.3923/ajbs.2024.796.808

This work is licensed under a Creative Commons Attribution 4.0 International License.


