Prevalence and Antimicrobial Susceptibility Testing and Analysis of Microbes Isolated from Different Clinical Samples
| Received 01 Apr, 2023 |
Accepted 15 Jul, 2023 |
Published 30 Sep, 2023 |
Background and Objective: In developing countries, the most crowded buildings in each city are hospitals and clinics due to the emergence of multidrug-resistant microbes. Therefore, the present study was conducted to find the resistance pattern of these microbes. Materials and Methods: A total of 2209 samples of blood, pus, tissue and urine were collected and cultured on their respective media plates. The culture-positive plates were identified through staining and biochemical methods. Antimicrobial susceptibility testing and statistical analysis, i.e., One-way ANOVA and post-hoc tests, were performed to check the resistance pattern and their significance. Results: Among cultured-positive samples, the most commonly isolated gram-positive cocci were Staphylococcus aureus and Enterococcus spp., which were resistant to Penicillin and Erythromycin and sensitive to Vancomycin and gram-negative rods were Escherichia coli and Klebsiella pneumoniae that were resistant to Ciprofloxacin and Ceftriaxone and sensitive to Polymyxin-B and Tigecycline. This study also witnessed the emergence of Serratia spp., as a multi-drug resistant bug and Proteus spp., as a secondary pathogen in skin and soft tissue infections. Conclusion: The antibiotics that are found to be highly resistant in this study should be stopped and microbial examination should be made compulsory before going for any treatment.
| Copyright © 2023 Naveed et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Bloodstream infection (BSI) fluctuates from acute infection to fatal sepsis, needs very fast antimicrobial care and has been considered a significant health concern. It causes clinical illness when viable microorganisms are present in the bloodstream1,2. The BSI are the most causative agents of morbidity and mortality in people of all age groups, especially in immune-compromised and hospitalized patients. About 30 million people are affected by BSI, which causes 6 million deaths globally, including 3 million newborns and 12 million children. Clinically important bacteria, including both Gram-negative rods (GNRs) and Gram-positive cocci (GPC), can cause BSI. The BSI treatment is typically based on the prevalent microbes and their antimicrobial susceptibilities. This information makes the basic recommendation for empirical therapy when BSI is detected3-5. A study was conducted by Al-Asady et al.6 to determine the incidence of sepsis and multidrug-resistant pathogens causing septicemia. The identified pathogens were S. aureus and S. hominis as the most common gram-positive bacteria and Salmonella typhi and S. gallinarum as the most common gram-negative bacteria. The gram-positive bacteria collectively were found highly resistant to Erythromycin, Clindamycin, Oxacillin, etc. and sensitive to Rifampicin, Moxifloxacin and Vancomycin while gram-negative bacteria were found highly resistant to Ceftazidime, Gentamicin and Amikacin and sensitive to Meropenem, Minocycline and Imipenem.
Skin and Soft Tissue Infections (SSTIs) involve the invasion of microorganisms into the skin and the underlying soft tissues (skin, fascia layer, subcutaneous fat and muscle-tendon structure) and can range from mild to critical infections and are categorized as uncomplicated or complicated7,8. The SSTIs are causing a high incidence rate in the US and worldwide. Due to the increased incidence of SSTIs in US, a study was conducted by Miller et al.9. Out of 48 million, 2,301,803 were identified as being infected with SSTIs. In these, 95% of SSTIs were treated in an ambulatory setting and 60% were categorized as abscesses or cellulitis. Different microorganisms are involved in causing different types of SSTIs10. A study was conducted to identify the microbiological profile of SSTIs from Northern India by Agrawal et al.11. The organisms isolated were Staphylococcus aureus, Pseudomonas aeruginosa, Klebsiella pneumoniae and the Acinetobacter baumannii complex. Among Coagulase-negative Staphylococcus are Staphylococcus hemolyticus, S. epidermidis, S. hominis and others. Among Enterococcus are Enterococcus faecium and E. faecalis. Antimicrobial drugs used to treat SSTIs depend on the microorganisms causing the infection and the type of infection11,12. Khan et al.13, conducted a study on 200 wound samples from Khyber Teaching Hospital (KTH) Peshawar for antimicrobial susceptibility testing. According to susceptibility profile, S. aureus was highly resistant to Amoxicillin, Oxafloxacin and Sparfloxacin etc. The gram-negative isolates were resistant to Cephalosporins, Augmentin and Quinolones. These pathogens were found resistant to Cefoperazone+Sulbactam, Carbapenems and Aminoglycosides.
Another important category of infection, the Urinary tract infections (UTIs) are the most common bacterial infections found in the female population. The UTIs can be complicated or uncomplicated and can cause a burden on the economy of a country14. The UTIs pose a serious threat to public health and are caused by a number of bacterial pathogens, such as E. coli, P. mirabilis, K. pneumoniae, E. faecalis and S. saprophyticus. Escherichia coli, Proteus spp. and Klebsiella spp., were isolated in a study which was conducted at the Regional Laboratory from 2003 to 2010 on 1404 urine samples. They were found resistant to Erythromycin, Amoxycillin and Tetracycline and sensitive to Nitrofurantoin, Ciprofloxacin and Gentamicin15,16.
The known multidrug-resistant bacterial pathogens that can cause clinical infections are MRSA, Enterococcus, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa and E. coli. They are referred to as ESKAPE pathogens17. Bacteria may be intrinsically resistant to some types of antibiotics or may acquire resistance through mutation (i.e., de novo mutation) or may acquire resistance genes from other organisms. The most dangerous form of antimicrobial resistance is the emergence or production of a “superbug” i.e., a pathogen that is resistant to many antibiotics or that is multidrug-resistant. In bacteria, multidrug resistance occurs either by the accumulation of genes or by the action of multidrug efflux pumps18,19.
The aim of this study was to identify the prevalence of infectious diseases in clinical settings and the pathogens that are causing such infections. Additionally, to find out the behavior of the same microbes isolated from different samples for the same set of antibiotics and the adverse effects of irrational use of broad-spectrum antibiotics through statistical analysis in different dimensions.
MATERIALS AND METHODS
Study design: This study was conducted at the Microbiology Laboratory in Tertiary Care Hospital, Peshawar, from 1st October, 2021 to 31st December, 2021.
Isolation of microbes: In this study, different samples (a total of 2209) were collected, including blood, pus, tissue and urine. Blood was drawn from the patients using sterile syringes (5 mL) and was immediately transferred to clean blood culture bottles. Pus samples were taken on swab sticks by rubbing them on the infected area. Tissue samples were taken from an infected tissue through biopsy techniques and that tissue was then kept in a sterile syringe or container depending on the size of the tissue and urine samples were taken in a sterile container. These samples were then cultured on their respective culture media plates. The blood, pus and tissue samples were cultured on Blood Agar and MacConkey Agar by the streak-plate method and urine samples were cultured on Cystine Lactose-Electrolyte-Deficient (CLED) media using a bacteriuria strip20. These plates were then incubated at 37°C for 12 to 24 hrs for pus, tissue and urine samples and 7 days for blood samples. The growth was retrieved the next day or the day after for pus, tissue and urine samples and on days 3-5 for blood samples.
Identification of bacteria: The samples that produced growth on culture media were then passed through different staining and biochemical techniques to identify the bacteria. Through gram staining, the isolated bacteria were examined microscopically to identify their morphology. The bacteria in this study were identified as gram-positive cocci (GPCs) and gram-negative rods (GNRs) on the basis of the color they retained and the shape they exhibited under the microscope. Different biochemical tests were then performed on GPCs and GNRs to identify their specific genus and species. Biochemical tests used for GPCs were the Catalase test to differentiate between Staphylococcus or Streptococcus bacteria and the Coagulase test (if catalase gave positive results) to differentiate S. aureus from S. saprophyticus and S. epidermidis. For GNRs, an oxidase test was performed to differentiate between Pseudomonadaceae and the Enterobacteriaceae family. The analytical profile index (API) test was then performed using API 10S strips to identify members of Enterobacteriaceae up to species level. Some colony characteristics on cultured media were also noted for accurate identification of isolates, such as size, color, margin, odor and elevation of colonies. The selective and differential media used in this study also provided an important area of identification, such as the hemolytic character of some microbes on blood agar and the production of pink-colored colonies of lactose fermenters on MacConkey agar.
Antimicrobial susceptibility testing: Antimicrobial susceptibility testing (AST) is pathology lab testing that determines the effective therapy of antibiotics against toxic and pathogenic bacteria. The AST was performed to find the culture sensitivity of microbes against conventional antibiotics using the the Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) AST standards. The disk diffusion method was used to perform AST21. It is one of the oldest approaches to AST and the most widely used method. This method is suitable for testing the majority of bacterial pathogens. Almost all antimicrobial agents can be tested and it doesn’t require any special equipment. The list of antibiotics used along with their abbreviation and disk potency are shown in Appendix A (Table A2).
Statistical analysis: Statistical analysis was performed using SPSS version 20. One-way ANOVA along with the homogeneity of variance test, Welch’s test and post-hoc test were performed to analyze the data statistically. One-way ANOVA was performed on GPCs and GNRs within each sample, microbes isolated from different samples and antibiotics used as a broad-spectrum treatment for different groups of microbes. The ANOVA test was also performed on MS Excel 2016 to compare it with the results of SPSS. Tests of homogeneity of variance and Welch’s test were performed to check the significance of ANOVA results. A post-hoc test was performed to identify the group that generated a different mean than the others. The Tukey’s HSD test was used as a first preference test, but the LSD test was also used as an alternative post-hoc test when the Tukey’s test didn’t give results. The null hypothesis for One-way ANOVA was “there is no difference between the means of groups” and the alternative hypothesis was “there is a difference between the means of groups” with a significance (p) value of 0.05 or less. The aim of this analysis was to check the significance of different resistance patterns shown by microbes in different samples for different antibiotics and to identify the microbe or antibiotic that is behaving differently for the same set of antibiotics or microbes, respectively.
| Table A1: | List of abbreviations | |||
| Abbreviation | Name |
| ABSSSIs | Acute Bacterial Skin and Soft Structure Infections |
| ANOVA | Analysis of Variance |
| API | Analytical Profile Index |
| AST | Antimicrobial Susceptibility Testing |
| BSI | Blood Stream Infections |
| CLED | Cystine Lactose-Electrolyte-Deficient |
| CLSI | Clinical and Laboratory Standards Institute |
| CONS | Coagulase Negative Staphylococci |
| ESKAPE | Enterococcus, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, E. coli |
| EUCAST | European Committee on Antimicrobial Susceptibility Testing |
| GNRs | Gram-Negative Rods |
| GPCs | Gram-Positive Cocci |
| HGT | Horizontal Gene Transfer |
| KP | Khyber Pakhtunkhwa |
| MRSA | Methicillin-Resistant Staphylococcus aureus |
| PCR | Polymerase Chain Reaction |
| SPSS | Statistical Package for Social Sciences |
| SSTIs | Skin and Soft Tissue Infections |
| UTIs | Urinary Tract Infections |
| Table A2: | List of antibiotics along with their disk potency and abbreviation | |||
| Sr. No. | Name of antibiotic | Potency (μg) |
Abbreviation | Sr. No. |
Name of antibiotic | Potency (μg) |
Abbreviation |
| 1 | Amikacin | 30 |
AK | 17 |
Fusidic acid | 10 |
FD |
| 2 | Ampicillin | 10 |
AMP | 18 |
Gentamicin | 10 |
GEN/CN |
| 3 | Azithromycin | 30 |
AZM | 19 |
Imipenem | 10 |
IMP/IPM |
| 4 | Aztreonam | 30 |
AZT/ATM | 20 |
Levofloxacin | 5 |
LEV |
| 5 | Cefepime | 30 |
FEP | 21 |
Linezolid | 30 |
LNZ/LDZ |
| 6 | Ceftazidime | 30 |
CAZ | 22 |
Meropenem | 10 |
MEM |
| 7 | Ceftriaxone | 30 |
CRO | 23 |
Nitrofurantoin | - |
NIT |
| 8 | Chloramphenicol | 30 |
CAP/C | 24 |
Penicillin | 10 |
P |
| 9 | Ciprofloxacin | 5 |
CIP | 25 |
Polymyxin-B | 300 |
PB |
| 10 | Clindamycin | 2 |
CLI | 26 |
Rifampicin | 5 |
RIF/RD |
| 11 | Cloxacillin | 30 |
CLOX | 27 |
Sulbactum-cefoperazone | 105 |
SCF |
| 12 | Co-amoxiclav | 30 |
AMC | 28 |
Tazobactum-peperacillin | 110 |
TZP |
| 13 | Co-trimoxazole | 25 |
COT | 29 |
Tigecycline | 15 |
TGC |
| 14 | Doxycycline | 30 |
DO/DOX | 30 |
Tobramycin | 10 |
TOB |
| 15 | Erythromycin | 15 |
ERY/E | 31 |
Vancomycin | 30 |
VAN |
| 16 | Fosfomycin | - |
FOS |
RESULTS
Isolation of microbes: This study was conducted at Tertiary Care Hospital, Peshawar. A total of 2209 clinical samples, of which 43.8% (n = 968) were blood samples, 14.9% (n = 330) were pus samples, 9.8% (n = 217) were tissue samples and 31.4% (n = 694) were urine samples, collected from 1st October, 2021 to 31st December, 2021. Out of these 2209 samples, only 21.7% (n = 479) were cultured positive, of which 17.3% (n = 83) were blood samples, 45.1% (n = 216) were pus samples, 21.7% (n = 104) were tissue samples and 15.9% (n = 76) were urine samples. Out of 479 cultured positive samples, 11.7% (n = 56) of the samples (pus and tissue samples) produced two different isolates. The total number of bacterial isolates in this study was 535. The age group and male to female ratio of samples were different for each sample. Table 1 represents the total number of samples, cultured positive samples, age group and gender-wise distribution of each sample.
|
| Table 1: | Total number of samples and age and gender wise distribution of samples | |||
Age groups and gender of patients with higher number of culture positive samples |
||||||
| Total number | Culture | Polymicrobial | Total number | |||
| Samples | of samples | positive | samples | of isolates | Gender | Age group |
| Blood | 968 | 83 | 0 | 83 | Male | 18 days-40 years |
| Pus | 330 | 216 | 27 | 243 | Male | 21-60 years |
| Tissue | 217 | 104 | 29 | 133 | Male | 21-60 years |
| Urine | 694 | 76 | 0 | 76 | Both male and female | 61-80 years |
| Total | 2209 | 479 | 56 | 535 | – |
Table 1 shows that blood and urine samples produce the greatest number of samples but the high number of culture-positive samples were produced by pus and tissue samples, which means that pus and tissue samples isolated from the sites are more susceptible to microbial invasion.
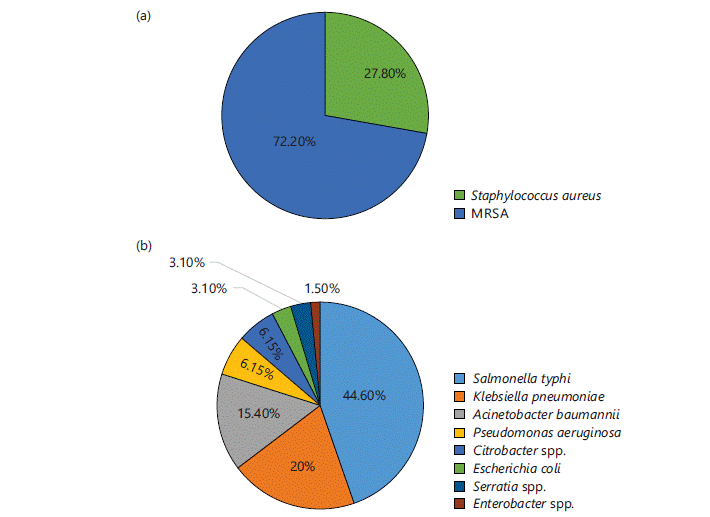
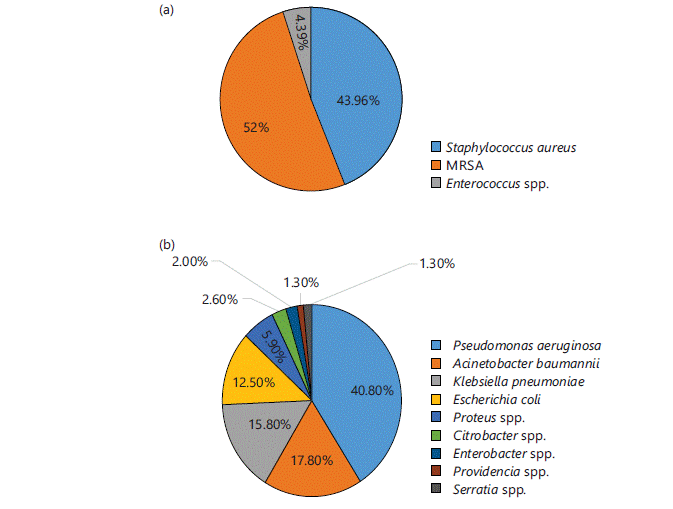
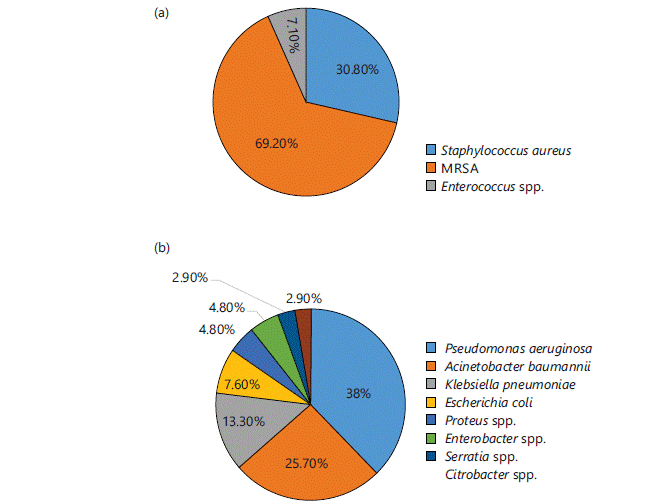
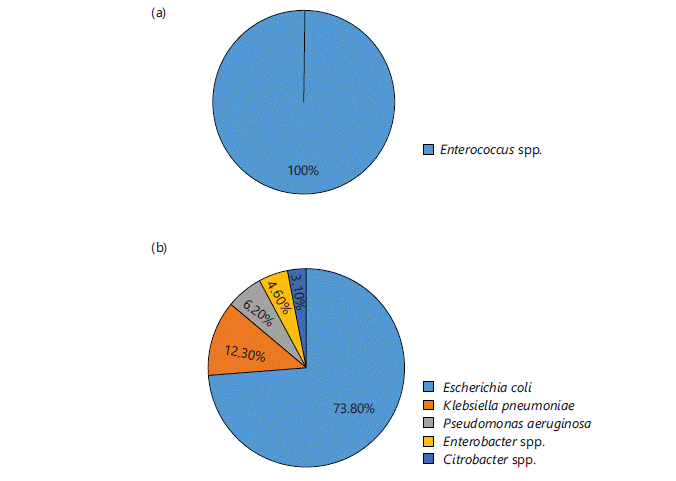
Identification of microbes: Different microbes were identified in different samples in different ratios. Out of 535 total isolates 27.7% (n = 148) were identified as GPCs and 72.3% (n = 387) were identified as GNRs. The microbes commonly isolated from all samples were S. aureus and MRSA (except urine samples) among GPCs and Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumannii, Citrobacter spp. and Enterobacter spp., among GNRs. Figure 1-4(a-b) represent different isolates with their percentages from different samples.
|
|
|
Figure 1 shows the percentage of isolated GPCs (Fig. 1a) and GNRs (Fig. 1b) in blood samples. Out of 83 culture-positive samples, 21.7% (n = 18) were identified as GPCs and 78.3% (n = 65) were identified as GNRs. The GPCs identified were S. aureus, of which 72.2% (n = 13) were Methicillin-resistant S. aureus (MRSA). Among GNRs, the highly isolated microbe in the blood sample was S. typhi: 44.6% (n = 29), followed by K. pneumoniae: 20% (n = 13), A. baumannii: 15.4% (n = 10), P. aeruginosa and Citrobacter spp.: 6.15% (n = 4), E. coli: 3.1% (n = 2), Serratia spp.: 3.1% (n = 2) and Enterobacter spp.: 1.5% (n = 1).
Figure 2 shows the percentage of isolated GPCs (Fig. 2a) and GNRs (Fig. 2b) from pus samples. Out of the total 243 isolates, 37.4% (n = 91) were GPCs and 62.5% (n = 152) were GNRs. Among the GPCs, 95.6% (n = 87) were identified as S. aureus, (of which half i.e., 54% (n = 47) were MRSA) and 4.4% (n = 4) were identified as Enterococcus spp. Among GNRs, 40.8% (n = 62) were P. aeruginosa, followed by 17.80% (n = 27) A. baumannii, 15.8% (n = 24) K. pneumoniae, 12.5% (n = 19) E. coli, 5.9% (n = 9) Proteus spp., 2.6% (n = 4) Citrobacter spp., 2.0% (n = 3) Enterobacter spp., 1.3% (n = 2) Providencia spp. and Serratia spp.
Figure 3 shows the percentage of isolated GPCs (Fig. 3a) and GNRs (Fig. 3b) from tissue samples. Out of total 133 isolates, 21.1% (n = 28) were identified as GPCs and 78.9% (n = 105) were identified as GNRs. Among GPCs, 92.9% (n = 26) were S. aureus (in which more than half, i.e., 69.2% (n = 18), were MRSA) and 7.10% (n = 2) were Enterococcus spp. Among GNRs, 38% (n = 40) were P. aeruginosa followed by 25.7% (n = 27) A. baumannii, 13.30% (n = 14) K. pneumoniae, 7.60% (n = 8) E. coli, 4.80% (n = 5) Proteus spp. and Enterobacter spp., 2.90% (n = 3) Serratia spp. and Citrobacter spp.
Figure 4 shows the percentage of isolated GPCs (Fig. 4a) and GNRs (Fig. 4b) in Urine samples. Out of total 76 isolates, 14.5% (n = 11) were GPCs, which were identified as Enterococcus spp. and 85.5% (n = 65) were GNRs, of which 73.80% (n = 48) were E. coli, followed by 12.30% (n = 8) K. pneumoniae, 6.20% (n = 4) P. aeruginosa, 4.60% (n = 3) Enterobacter spp. and 3.1% (n = 2) Citrobacter spp.
Antimicrobial susceptibility testing: The identified microbes from each sample were tested for antibiotic susceptibility against different antibiotics used for different groups of species. Table 2-5 shows the antibiotic susceptibility pattern of microbes for each sample. A different set of antibiotics was used for GPCs and GNRs according to their cellular structures and modes of action of antibiotics. Among GPCs, Enterococcus spp. and among GNRs, P. aeruginosa and S. typhi were treated with antibiotics other than those used for typical GPCs and GNRs.
Table 2: |
Antibiotic susceptibility testing of microbes isolated from blood samples |
|
R: Resistant, S: Sensitive, IR: Intrinsically resistant, N: Not used (i.e., this antibiotic was not used for this microbe), -: 0-25%, +: 26-50%, ++: 51-75%, +++: 76-100% and *Changes in percentages were due to unavailability of certain resources |
||
Table 2 shows the antibiotic susceptibility testing or culture sensitivity of microbes isolated from blood samples. From this table, it is clear that GPCs, i.e., S. aureus and MRSA collectively, were highly resistant to P (100%) followed by ERY (83.3%) and COT (61.6%) and were highly sensitive to CLOX and AMC (100%, except MRSA, which was intrinsically resistant for them) and LNZ and VAN (100%) followed by RIF and GEN (83.3%), DOX (77.8%), CLI (72.2%), CIP (61.1%) and FD (55.6%). Salmonella typhi was found highly resistant to AMP and CIP (93.1%), followed by CRO (79.3%) and COT (55.1%) and highly sensitive to AZM (89.7%), followed by MEM (82.8%) and IMP (48.3%). Pseudomonas aeruginosa was found highly resistant to CAZ (100%) and highly sensitive to TZP, CIP, IMP, MEM and PB (100%), followed by GEN, TOB, AZM, FEP and AK (75%). Other GNRs, i.e., K. pneumoniae, A. baumannii, Citrobacter spp., E. coli, Serratia spp. and Enterobacter spp., collectively were highly resistant to AZM (77.3%, except A. baumannii, which was intrinsically resistant to it), followed by CRO (68.8%), CIP and MEM (65.6%), COT (56.2%) and SCF (50%) and were highly sensitive to TGC (93.8%), followed by PB (93.3%, except Serratia spp., which was intrinsically resistant to PB), GEN (59.4%), DOX and TZP (56.2%) and AK (50%).
Table 3 shows the antibiotic susceptibility testing or culture sensitivity of microbes isolated from pus samples. As it is clear from the table, among GPCs, S. aureus and MRSA collectively were highly resistant to P (85%), followed by CIP (68.96%) and ERY (67.8%) and highly sensitive to CLOX and AMC (100%, except MRSA), followed by VAN (98.9%), LNZ (97.7%), RIF (96.6%), DOX (90.8%), CLI and GEN (79.3%), COT (75.9%) and FD (71.3%). Enterococcus spp., was found to be highly resistant to ERY (100%) followed by P and DOX (75%) and highly sensitive to VAN (100%) followed by CAP (75%). Among GNRs, P. aeruginosa was highly resistant to CAZ (88.7%), followed by MEM (70.97%), CIP (69.4%), TOB and FEP (66.1%), IMP (64.5%), GEN and AK (58.1%), TZP (56.5%) and AZT (50%) and highly sensitive to PB (100%). Other GNRs i.e., A. baumannii, K. pneumoniae, E. coli, Proteus spp., Citrobacter spp., Enterobacter spp., Providencia spp. and Serratia spp., collectively were highly resistant to CIP (76.7%) followed by CRO (71.1%), FEP and COT (68.9%), TZP (58.9%), GEN (53.3%) and MEM (52.2%) and highly sensitive to PB (100%, except Proteus spp., Providencia spp. and Serratia spp., which were intrinsically resistant to this) followed by TGC (98.7%, without Proteus spp. and Providencia spp.), CAP (63.5%), AK (61.1%), DOX (53.4%) and IMP and SCF (51.5%).
Table 4 represents the antibiotic susceptibility testing or culture sensitivity of microbes isolated from tissue samples, which shows that among GPCs, S. aureus and MRSA collectively were found highly resistant to P (100%) followed by ERY (76.9%) and CIP (73.7%) and highly sensitive to VAN (100%) followed by RIF and LNZ (96.2%), DOX (92.3%), CLI (88.5%), GEN (80.8%), CLOX and AMC (75%, except MRSA), FD (73.1%) and COT (69.2%). Enterococcus spp., was highly resistant to ERY (100%) and highly sensitive to LNZ and VAN (100%), while P and DOX were 50% resistant and 50% sensitive. Among GNRs, P. aeruginosa was found highly resistant to CAZ (92.5%), followed by TOB (85%), CIP (82.5%), AZT (72.5%), GEN and RIF (70%), MEM and AK (67.5%) and IMP (65%) and highly sensitive to PB (100%) followed by TZP (47.5%). Other GNRs, i.e., A. baumannii, K. pneumoniae, E. coli, Proteus spp., Enterobacter spp., Serratia spp. and Citrobacter spp., collectively, were found highly resistant to CRO (84.6%), followed by CIP (83.1%), COT (75.4%), FEP (73.8%), TZP (70.8%), GEN (67.7%), MEM (61.5%), AK (58.5%), IMP (56.9%), SCF (55.4%) and AZT (52.6%, except A. baumannii) and highly sensitive to PB (100%, except Proteus spp. and Serratia spp.), followed by TGC (93.3%, except Serratia spp.) and CAP (57.9%).
Table 3: |
Antibiotic susceptibility of microbes isolated from pus samples |
|
R: Resistant, S: Sensitive, IR: Intrinsically resistant, N: Not used (i.e., this antibiotic was not used for this microbe), -: 0-25%, +: 26-50%, ++: 51-75%, +++: 76-100% and *Changes in percentages were due to unavailability of certain resources |
||
Table 4: |
Antibiotic susceptibility testing of microbes isolated from tissue samples |
|
R: Resistant, S: Sensitive, IR: Intrinsically resistant, N: Not used (i.e., this antibiotic was not used for this microbe), -: 0-25%, +: 26-50%, ++: 51-75%, +++: 76-100% and *Changes in percentages were due to unavailability of certain resources |
||
Table 5: |
Antibiotic susceptibility testing of microbes isolated from urine samples |
|
R: Resistant, S: Sensitive, IR: Intrinsically resistant, N: Not used (i.e., this antibiotic was not used for this microbe), -: 0-25%, +: 26-50%, ++: 51-75%, +++: 76-100% and *Changes in percentages were due to unavailability of certain resources |
||
| Table 6: | Data of blood sample for statistical analysis | |||
Blood |
|||||||||||
GPCs |
GNRs |
||||||||||
| Antibiotics | S. aureus |
MRSA |
Antibiotics |
P. aeruginosa |
Antibiotics |
K. pneumoniae |
A. baumannii |
Citrobacter spp. |
E. coli |
Serratia spp. |
Enterobacter spp. |
| P | 1 |
1 |
CAZ |
1 |
AMP |
1 |
1 |
1 |
0.5 |
1 |
1 |
| CLOX | 0 |
1 |
GEN |
0.25 |
AMC |
0.08 |
1 |
1 |
1 |
1 |
1 |
| AMC | 0 |
1 |
TZP |
0 |
GEN |
0.54 |
0.3 |
0.25 |
0.5 |
0.5 |
0 |
| CIP | 0.2 |
0.46 |
CIP |
0 |
CIP |
0.85 |
0.5 |
0.5 |
0.5 |
0.5 |
1 |
| ERY | 0.6 |
0.92 |
TOB |
0.25 |
CRO |
0.85 |
0.6 |
0.25 |
1 |
0.5 |
1 |
| COT | 0.6 |
0.61 |
IMP |
0 |
FEP |
0.77 |
0.6 |
0.25 |
1 |
0.5 |
1 |
| CLI | 0 |
0.38 |
MEM |
0 |
DOX |
0.46 |
0.1 |
0 |
1 |
1 |
1 |
| DOX | 0.4 |
0.15 |
AZT |
0.25 |
TZP |
0.69 |
0.2 |
0.25 |
0.5 |
0 |
1 |
| RIF | 0 |
0.07 |
FEP |
0.25 |
COT |
0.62 |
0.4 |
0.5 |
1 |
0.5 |
1 |
| LNZ | 0 |
0 |
AK |
0.25 |
AK |
0.62 |
0.4 |
0.5 |
0 |
0.5 |
0 |
| GEN | 0 |
0.23 |
PB |
0 |
IMP |
0.54 |
0.2 |
0.25 |
0.5 |
0 |
1 |
| FD | 0.6 |
0.38 |
MEM |
0.77 |
0.5 |
0.75 |
0.5 |
0.5 |
1 |
||
| VAN | 0 |
0 |
CAP |
0.46 |
1 |
0.25 |
0.5 |
0.5 |
0 |
||
| CAP | 0 |
0 |
AZM |
0.77 |
1 |
0.5 |
1 |
1 |
1 |
||
| TGC | 0 |
0 |
SCF |
0.69 |
0.3 |
0.25 |
0.5 |
0.5 |
1 |
||
TGC |
0 |
0 |
0 |
0.5 |
0 |
0 |
|||||
PB |
0 |
0 |
0.5 |
0 |
1 |
0 |
|||||
| AK: Amikacin, AMP: Ampicillin, AZM: Azithromycin, AZT: Aztreonam, FEP: Cefepime, CAZ: Ceftazidime, CRO: Ceftriaxone, CAP: Chloramphenicol, CIP: Ciprofloxacin, CLI: Clindamycin, CLOX: Cloxacillin, AMC: Co-amoxiclav, COT: Co-trimoxazole, DOX: Doxycycline, ERY: Erythromycin, FD: Fusidic acid, GEN: Gentamicin, IMP: Imipenem, LNZ: Linezolid, MEM: Meropenem, P: Penicillin, PB: Polymyxin-B, RIF: Rifampicin, SCF: Sulbactum-Cefoperazone, TZP: Tazobactum-Peperacillin, TGC: Tigecycline, TOB: Tobramycin, and VAN: Vancomycin | |||||||||||
Table 5 shows the antibiotic susceptibility testing or culture sensitivity of microbes isolated from urine samples. From the table, it is clear that among GPCs, Enterococcus spp., was highly resistant to LEV, FOS and CIP (63.7%) and sensitive to LNZ and VAN (100%), followed by NIT (81.8%), P and ERY (54.5%). Among GNRs, P. aeruginosa was found highly sensitive to CAZ (100%) and sensitive to PB (100%), followed by AK (75%), while for others it was 50% resistant and 50% sensitive. Other GNRs, i.e., E. coli, K. pneumoniae, Enterobacter spp. and Citrobacter spp., collectively, were highly resistant to CIP (78.7%) followed by CRO (72.1%), FEP (65.6%) and COT (63.9%) and were highly sensitive to IMP (85.2%), followed by AK and MEM (83.6%), SCF (75.4%), CAP (72.1%), TZP (65.6%) and GEN (62.3%).
Statistical analysis: Table 6-9 shows the data of each sample for statistical analysis. The data was taken in the form of 0 to 1, i.e., the mean resistance value of each microbe for each antibiotic, e.g. if a microbe is 100% resistant to an antibiotic, then its mean value will be 1 and if it is 50% resistant, then its mean value will be 0.5. This is to avoid errors due to the sample size of each microbe within a sample and in different samples. This data was used in different combinations to find the reason for different resistant patterns of different microbes in different samples for same antibiotics. Note that for certain analysis’ the assumptions of homogeneity or homogeneity of variances were violated due to different sample sizes or the occurrence of any discontinuous data, so therefore the significance value of Welch’s test was used as an alternative for these analysis’. One-way ANOVA was also performed on MS Excel 2016 to compare its results with SPSS results for confirmation.
Statistical analysis of GPCs and GNRs within each sample: A One-way ANOVA along with a post-hoc Tukey’s test was first performed on the GPCs and GNRs of each sample individually to find out whether the different groups of GPCs and GNRs for which the same set of antibiotics were used are statistically significant or not, whether the means are the same or not, or whether they behave the same or not.
On GPCs and GNRs in blood samples: The GPCs and GNRs isolated from blood samples behaved the same for the set of antibiotics used and their means of resistance were also the same. It could be verified from the ANOVA results of GPCs and GNRs in blood samples from Table 10 and 11. From Table 10, the significance value is 0.173, which is greater than 0.05, so the null hypothesis will be accepted, which states that the means of the groups are the same and from Table 11, the sig. value is 0.209. which is again greater than 0.05, so the null hypothesis will be accepted in this case too.
On GPCs and GNRs in pus sample: In pus samples, GPCs, i.e., S. aureus and MRSA, show the same pattern of resistance and sensitivity against the same set of antibiotics, as is evident from Table 12, which shows the One-way ANOVA results of GPCs isolated from pus samples. Its significance value is 0.173, which is greater than 0.05, so the null hypothesis will be accepted that there is no difference between the means of the groups. On the other hand, the GNRs of the pus sample didn’t follow the same pattern. From Table 13a, which shows the results of a One-way ANOVA of GNRs isolated from pus samples, the significance value is 0.000 (from MS Excel ANOVA results, its 0.0000429), which is way too smaller than 0.05, so the alternative hypothesis will be accepted, which states that there is a difference in the means of any one or more groups analyzed. So, to find out the group that behaved differently, the Tukey’s test was performed as a post-hoc test, which is shown in Table 13b.
So it is clear from Table 13b and the descriptive table (which is not shown here) that Serratia spp., was behaving differently than all these groups, with a mean value much greater than the others. This is because Serratia spp., show different patterns of resistance to certain antibiotics that will be discussed later in the statistical analysis on microbes and antibiotics section. And also, some antibiotics were intrinsically resistant for some groups but sensitive or slightly resistant for others. So it can also cause variations in their means. Note that the homogeneity of variance was violated in the analysis of GNRs in pus, so as an alternative, the result of Welch’s test was used, whose significance value was also 0.000.
Table 7: |
Data of pus sample for statistical analysis |
|
AK: Amikacin, AMP: Ampicillin, AZM: Azithromycin, AZT: Aztreonam, FEP: Cefepime, CAZ: Ceftazidime, CRO: Ceftriaxone, CAP: Chloramphenicol, CIP: Ciprofloxacin, CLI: Clindamycin, CLOX: Cloxacillin, AMC: Co-amoxiclav, COT: Co-trimoxazole, DOX: Doxycycline, ERY: Erythromycin, FD: Fusidic acid, GEN: Gentamicin, IMP: Imipenem, LNZ: Linezolid, MEM: Meropenem, P: Penicillin, PB: Polymyxin-B, RIF: Rifampicin, SCF: Sulbactum-Cefoperazone, TZP: Tazobactum-Peperacillin, TGC: Tigecycline, TOB: Tobramycin and VAN: Vancomycin |
||
| Table 8: | Data of tissue sample for statistical analysis | |||
Tissue |
||||||||||||||
GPCs |
GNRs |
|||||||||||||
| Antibiotics | S. aureus |
MRSA |
Antibiotics |
Enterococcus spp., |
Antibiotics |
P. aeruginosa |
Antibiotics |
A. baumannii |
K. pneumoniae |
E. coli |
Proteus spp., |
Enterobacter spp., |
Serratia spp., |
Citrobacter spp., |
| P | 1 |
1 |
P |
0.5 |
CAZ |
0.93 |
AMP |
1 |
1 |
0.88 |
0.8 |
1 |
1 |
1 |
| CLOX | 0.25 |
1 |
ERY |
1 |
GEN |
0.7 |
AMC |
1 |
0.36 |
0.38 |
0.6 |
1 |
1 |
1 |
| AMC | 0.25 |
1 |
DOX |
0.5 |
TZP |
0.5 |
GEN |
0.67 |
0.79 |
0.75 |
0.6 |
0.6 |
0 |
1 |
| CIP | 0.88 |
0.67 |
RIF |
0 |
CIP |
0.83 |
CIP |
0.89 |
0.86 |
0.63 |
1 |
0.6 |
1 |
0.67 |
| ERY | 0.5 |
0.89 |
LNZ |
0 |
TOB |
0.85 |
CRO |
1 |
0.93 |
0.63 |
0.4 |
0.6 |
0.67 |
1 |
| COT | 0.13 |
0.39 |
VAN |
0 |
IMP |
0.65 |
FEP |
0.89 |
0.79 |
0.5 |
0.4 |
0.4 |
0.67 |
1 |
| CLI | 0.13 |
0.11 |
CAP |
0 |
MEM |
0.68 |
DOX |
0.26 |
0.57 |
0.5 |
0.4 |
0.6 |
0.67 |
0.67 |
| DOX | 0.13 |
0.06 |
AMP |
0 |
AZT |
0.73 |
TZP |
0.93 |
0.71 |
0.5 |
0 |
0.4 |
0.67 |
1 |
| RIF | 0.13 |
0 |
FEP |
0.7 |
COT |
0.67 |
1 |
0.63 |
0.8 |
0.6 |
0.67 |
1 |
||
| LNZ | 0.13 |
0 |
AK |
0.68 |
AK |
0.78 |
0.64 |
0 |
0.4 |
0.2 |
0.67 |
1 |
||
| GEN | 0.13 |
0.22 |
PB |
0 |
IMP |
0.93 |
0.5 |
0.13 |
0 |
0.4 |
0.33 |
0.33 |
||
| FD | 0.25 |
0.28 |
MEM |
0.93 |
0.64 |
0.25 |
0 |
0.4 |
0 |
0.67 |
||||
| VAN | 0 |
0 |
CAP |
1 |
0.36 |
0.38 |
0.6 |
0.4 |
0.33 |
0 |
||||
| CAP | 0 |
0 |
AZT |
1 |
0.64 |
0.5 |
0.4 |
0 |
0.67 |
1 |
||||
| TGC | 0 |
0 |
SCF |
0.67 |
0.71 |
0.38 |
0 |
0.4 |
0.67 |
0.33 |
||||
TGC |
0.04 |
0 |
0.13 |
1 |
0 |
0 |
0 |
|||||||
PB |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
|||||||
| AK: Amikacin, AMP: Ampicillin, AZT: Aztreonam, FEP: Cefepime, CAZ: Ceftazidime, CRO: Ceftriaxone, CAP: Chloramphenicol, CIP: Ciprofloxacin, CLI: Clindamycin, CLOX: Cloxacillin, AMC: Co-amoxiclav, COT: Co-trimoxazole, DOX: Doxycycline, ERY: Erythromycin, FD: Fusidic acid, GEN: Gentamicin, IMP: Imipenem, LNZ: Linezolid, MEM: Meropenem, P: Penicillin, PB: Polymyxin-B, RIF: Rifampicin, SCF: Sulbactum-Cefoperazone, TZP: Tazobactum-Peperacillin, TGC: Tigecycline, TOB: Tobramycin and VAN: Vancomycin | ||||||||||||||
| Table 9: | Data of urine sample for statistical analysis | |||
Urine |
||||||||
GPCs |
GNRs |
|||||||
| Antibiotics | Enterococcus spp. |
Antibiotics |
P. aeruginosa |
Antibiotics |
E. coli |
K. pneumoniae |
Enterobacter spp. |
Citrobacter spp. |
P | 0.36 |
CAZ |
1 |
AMP |
0.98 |
1 |
1 |
1 |
| AMC | 0.36 |
GEN |
0.5 |
AMC |
0.33 |
0.38 |
1 |
1 |
| CIP | 0.64 |
TZP |
0.5 |
GEN |
0.4 |
0.38 |
0 |
0.5 |
| LEV | 0.64 |
CIP |
0.5 |
CIP |
0.88 |
0.5 |
0.33 |
0.5 |
| LNZ | 0 |
TOB |
0.5 |
CRO |
0.79 |
0.5 |
0.33 |
0.5 |
| VAN | 0 |
IMP |
0.5 |
FEP |
0.71 |
0.5 |
0.33 |
0.5 |
| FOS | 0.64 |
MEM |
0.5 |
TZP |
0.33 |
0.38 |
0 |
0.5 |
| NIT | 0.18 |
AZT |
0.5 |
COT |
0.71 |
0.38 |
0.33 |
0.5 |
FEP |
0.5 |
AK |
0.13 |
0.38 |
0 |
0 |
||
AK |
0.25 |
IMP |
0.13 |
0.25 |
0 |
0.5 |
||
PB |
0 |
MEM |
0.13 |
0.38 |
0 |
0.5 |
||
SCF |
0.23 |
0.25 |
0 |
0.5 |
||||
CAP |
0.1 |
0 |
0 |
0.5 |
||||
| *AK: Amikacin, AMP: Ampicillin, AZT: Aztreonam, FEP: Cefepime, CAZ: Ceftazidime, CRO: Ceftriaxone, CAP: Chloramphenicol, CIP: Ciprofloxacin, AMC: Co-amoxiclav, COT: Co-trimoxazole, FOS: Fosfomycin, GEN: Gentamicin, IMP: Imipenem, LEV: Levofloxacin, LNZ: Linezolid, MEM: Meropenem, NIT: Nitrofurantoin, P: Penicillin, PB: Polymyxin-B, SCF: Sulbactum-Cefoperazone, TZP: Tazobactum-Peperacillin, TOB: Tobramycin and VAN: Vancomycin | ||||||||
| Table 10: | One-way ANOVA results of GPCs in blood sample | |||
ANOVA |
|||||
| Source of variation | Sum of square |
df |
Mean square |
F |
Significance |
| Between groups | 0.261 |
1 |
0.261 |
1.953 |
0.173 |
| Within groups | 3.746 |
28 |
0.134 |
||
| Total | 4.007 |
29 |
|||
| *F: F-statistic, a ratio of two variances that measure the dispersal of the data points around the mean and df: Degrees of freedom, the number of independent pieces of information | |||||
| Table 11: | One-way ANOVA results of GNRs in blood sample | |||
ANOVA |
|||||
| Source of variation | Sum of square |
df |
Mean square |
F |
Significance |
| Between groups | 0.912 |
5 |
0.182 |
1.463 |
0.209 |
| Within groups | 11.965 |
96 |
0.125 |
||
| Total | 12.877 |
101 |
|||
| *F: F-statistic, a ratio of two variances that measure the dispersal of the data points around the mean and df: Degrees of freedom, the number of independent pieces of information | |||||
| Table 12: | One-way ANOVA results of GPCs in pus sample | |||
ANOVA |
|||||
| Source of variation | Sum of square |
df |
Mean square |
F |
Significance |
| Between groups | 0.231 |
1 |
0.231 |
1.957 |
0.173 |
| Within groups | 3.3 |
28 |
0.118 |
||
| Total | 3.53 |
29 |
|||
| *F: F-statistic, a ratio of two variances that measure the dispersal of the data points around the mean and df: Degrees of freedom, the number of independent pieces of information | |||||
On GPCs and GNRs in tissue sample: From Table 14, it is clear that GPCs isolated from tissue samples show the same pattern of resistance for the same set of antibiotics used. The significance value from the ANOVA results is 0.399, which is greater than 0.05, so the null hypothesis will be accepted. While, the GNRs isolated from tissue samples generate different resistance patterns as shown in Table 15. Table 15a shows the ANOVA results, in which the significance value for GNRs is 0.036, which is smaller than 0.05, so by accepting the alternative hypothesis, it is concluded that the means of resistance pattern of GNRs are not the same. A post-hoc test was also performed to find out which group was different from others. But Tukey’s test didn’t give any significant results, so the LSD test was used as a post-hoc test in this case. Table 15b shows the results of LSD as a post-hoc test for GNRs in tissue samples. As it is clear from Table 15b that there are variations among different groups and also, after observing their mean values from the descriptive table (not shown here), any one group could not be targeted. Approximately all groups showed differences with one or more other groups. This is because the set of antibiotics used contains some antibiotics that are intrinsically resistant for some groups but sensitive for others, e.g., PB was intrinsically resistant to Proteus spp. and Serratia spp., but was 100% sensitive for all others.
| Table 13: | One-way ANOVA and post-hoc test results of GNRs in pus samples | |||
| (a) One-way ANOVA results of GNRs in pus sample | ||||||
ANOVA |
||||||
| Source of variation | Sum of squares |
df |
Mean square |
F |
Significance |
|
| Between groups | 4.144 |
7 |
0.592 |
5.061 |
0.000 |
|
| Within groups | 14.974 |
128 |
0.117 |
|||
| Total | 19.119 |
135 |
||||
| (b): Post-hoc test results of GNRs in the pus sample | ||||||
Tukey’s HSD |
||||||
95% Confidence interval |
||||||
| (I) Samples | (J) Samples | Mean difference (I-J) |
Std. Error |
Significance |
Lower bound |
Upper bound |
| A. baumannii | K. pneumoniae | 0.18294 |
0.11732 |
0.773 |
-0.1786 |
0.5445 |
| E. coli | 0.24824 |
0.11732 |
0.41 |
-0.1133 |
0.6098 |
|
| Proteus spp. | 0.30294 |
0.11732 |
0.172 |
-0.0586 |
0.6645 |
|
| Citrobacter spp. | 0.02706 |
0.11732 |
1 |
-0.3345 |
0.3886 |
|
| Enterobacter spp. | 0.23824 |
0.11732 |
0.466 |
-0.1233 |
0.5998 |
|
| Providencia spp. | 0.36529* |
0.11732 |
0.046 |
0.0038 |
0.7268 |
|
| Serratia spp. | -0.19353 |
0.11732 |
0.719 |
-0.5551 |
0.168 |
|
| K. pneumoniae | E. coli | 0.06529 |
0.11732 |
0.999 |
-0.2962 |
0.4268 |
| Proteus spp. | 0.12 |
0.11732 |
0.97 |
-0.2415 |
0.4815 |
|
| Citrobacter spp. | -0.15588 |
0.11732 |
0.886 |
-0.5174 |
0.2057 |
|
| Enterobacter spp. | 0.05529 |
0.11732 |
1 |
-0.3062 |
0.4168 |
|
| Providencia spp. | 0.18235 |
0.11732 |
0.776 |
-0.1792 |
0.5439 |
|
| Serratia spp. | -0.37647* |
0.11732 |
0.035 |
-0.738 |
-0.0149 |
|
| E. coli | Proteus spp. | 0.05471 |
0.11732 |
1 |
-0.3068 |
0.4162 |
| Citrobacter spp. | -0.22118 |
0.11732 |
0.563 |
-0.5827 |
0.1404 |
|
| Enterobacter spp. | -0.01 |
0.11732 |
1 |
-0.3715 |
0.3515 |
|
| Providencia spp. | 0.11706 |
0.11732 |
0.974 |
-0.2445 |
0.4786 |
|
| Serratia spp. | -0.44176* |
0.11732 |
0.006 |
-0.8033 |
-0.0802 |
|
| Proteus spp. | Citrobacter spp. | -0.27588 |
0.11732 |
0.275 |
-0.6374 |
0.0857 |
| Enterobacter spp. | -0.06471 |
0.11732 |
0.999 |
-0.4262 |
0.2968 |
|
| Providencia spp. | 0.06235 |
0.11732 |
0.999 |
-0.2992 |
0.4239 |
|
| Serratia spp. | -0.49647* |
0.11732 |
0.001 |
-0.858 |
-0.1349 |
|
| Citrobacter spp. | Enterobacter spp. | 0.21118 |
0.11732 |
0.621 |
-0.1504 |
0.5727 |
| Providencia spp. | 0.33824 |
0.11732 |
0.085 |
-0.0233 |
0.6998 |
|
| Serratia spp. | -0.22059 |
0.11732 |
0.567 |
-0.5821 |
0.1409 |
|
| Enterobacter spp. | Providencia spp. | 0.12706 |
0.11732 |
0.959 |
-0.2345 |
0.4886 |
| Serratia spp. | -0.43176* |
0.11732 |
0.008 |
-0.7933 |
-0.0702 |
|
| Providencia spp. | Serratia spp. | -0.55882* |
0.11732 |
0 |
-0.9204 |
-0.1973 |
| (a) *F: F-statistic, a ratio of two variances that measure the dispersal of the data points around the mean and df: Degrees of freedom, the number of independent pieces of information, (b) *Mean difference is significant at the 0.05 level and *Certain modifications were done in this table to avoid repetition | ||||||
| Table 14: | One-way ANOVA results of GPCs in tissue sample | |||
ANOVA |
|||||
| Source of variation | Sum of square |
df |
Mean square |
F |
Significance |
| Between groups | 0.097 |
1 |
0.097 |
0.733 |
0.399 |
| Within groups | 3.724 |
28 |
0.133 |
||
| Total | 3.822 |
29 |
|||
| *F: F-statistic, a ratio of two variances that measure the dispersal of the data points around the mean and df: Degrees of freedom, the number of independent pieces of information | |||||
| Table 15: | One-way ANOVA and post-hoc test results of GNRs in tissue sample | |||
| (a): One-way ANOVA results of GNRs in tissue sample | ||||||
ANOVA |
||||||
| Source of variation | Sum of square | df | Mean square | F | Significance | |
| Between groups | 1.523 | 6 | 0.254 | 2.346 | 0.036 | |
| Within groups | 12.115 | 112 | 0.108 | |||
| Total | 13.638 | 118 | ||||
| (b): Post-hoc test results of GNRs in tissue sample | ||||||
LSD |
||||||
| 95% Confidence interval | ||||||
| (I) Samples | (J) Samples | Mean difference (I-J) |
Std. Error |
Significance |
Lower bound |
Upper bound |
| A. baumannii | K. pneumoniae | 0.12706 |
0.11281 |
0.262 |
-0.0965 |
0.3506 |
| E. coli | 0.32294* |
0.11281 |
0.005 |
0.0994 |
0.5465 |
|
| Proteus spp. | 0.25059* |
0.11281 |
0.028 |
0.0271 |
0.4741 |
|
| Enterobacter spp. | 0.29765* |
0.11281 |
0.01 |
0.0741 |
0.5212 |
|
| Serratia spp. | 0.15529 |
0.11281 |
0.171 |
-0.0682 |
0.3788 |
|
| Citrobacter spp. | 0.05824 |
0.11281 |
0.607 |
-0.1653 |
0.2818 |
|
| K. pneumoniae | E. coli | 0.19588 |
0.11281 |
0.085 |
-0.0276 |
0.4194 |
| Proteus spp. | 0.12353 |
0.11281 |
0.276 |
-0.1 |
0.347 |
|
| Enterobacter spp. | 0.17059 |
0.11281 |
0.133 |
-0.0529 |
0.3941 |
|
| Serratia spp. | 0.02824 |
0.11281 |
0.803 |
-0.1953 |
0.2518 |
|
| Citrobacter spp. | -0.06882 |
0.11281 |
0.543 |
-0.2923 |
0.1547 |
|
| E. coli | Proteus spp. | -0.07235 |
0.11281 |
0.523 |
-0.2959 |
0.1512 |
| Enterobacter spp. | -0.02529 |
0.11281 |
0.823 |
-0.2488 |
0.1982 |
|
| Serratia spp. | -0.16765 |
0.11281 |
0.14 |
-0.3912 |
0.0559 |
|
| Citrobacter spp. | -0.26471* |
0.11281 |
0.021 |
-0.4882 |
-0.0412 |
|
| Proteus spp. | Enterobacter spp. | 0.04706 |
0.11281 |
0.677 |
-0.1765 |
0.2706 |
| Serratia spp. | -0.09529 |
0.11281 |
0.4 |
-0.3188 |
0.1282 |
|
| Citrobacter spp. | -0.19235 |
0.11281 |
0.091 |
-0.4159 |
0.0312 |
|
| Enterobacter spp. | Serratia spp. | -0.14235 |
0.11281 |
0.21 |
-0.3659 |
0.0812 |
| Citrobacter spp. | -0.23941* |
0.11281 |
0.036 |
-0.4629 |
-0.0159 |
|
| Serratia spp. | Citrobacter spp. | -0.09706 |
0.11281 |
0.391 |
-0.3206 |
0.1265 |
| (a) *F: F-statistic, a ratio of two variances that measure the dispersal of the data points around the mean and df: Degrees of freedom, the number of independent pieces of information and (b) *Mean difference is significant at the 0.05 level and *Certain modifications were done to avoid repetition | ||||||
| Table 16: | One-way ANOVA results of GNRs in urine sample | |||
ANOVA |
|||||
| Source of variation | Sum of square |
df |
Mean square |
F |
Significance |
| Between groups | 0.546 |
3 |
0.182 |
2.109 |
0.111 |
| Within groups | 4.141 |
48 |
0.086 |
||
| Total | 4.687 |
51 |
|||
| *F: F-statistic, a ratio of two variances that measure the dispersal of the data points around the mean and df: Degrees of freedom, the number of independent pieces of information | |||||
On GNRs in urine sample: The GPCs in the urine sample were not analyzed because it has only 1 group, i.e., Enterococcus spp. The GNRs isolated from the urine sample show the same resistance pattern as their means were measured the same on the One-way ANOVA. Table 16 shows the results of a One-way ANOVA of GNRs in urine samples. From Table 16, the significance value can be noted, which is 0.111, which is greater than 0.05, so the null hypothesis will be accepted that the means of groups of GNRs in the urine sample are the same.
| Table 17: | Statistical analysis of microbes isolated from different samples | |||
ANOVA results |
||||
| Microbe | Samples | F |
Significance |
post-hoc results (Tukey’s HSD) |
| S. aureus | Blood, pus and tissue | 1.35 |
3.22 |
Nil |
| MRSA | Blood, pus and tissue | 0.04 |
0.961 |
Nil |
| Enterococcus spp. | Pus, tissue and urine | 1 |
0.334 |
Nil |
| P. aeruginosa | All the four samples | 7.024 |
0.001 |
Blood-pus: 0.005 |
| Blood-tissue: 0.001 | ||||
| A. baumannii | Blood, pus and tissue | 2.929 |
0.063 |
Nil |
| K. pneumoniae | All the four samples | 3.843 |
0.014 |
Blood-urine: 0.046 |
| Tissue-urine: 0.013 | ||||
| E. coli | All the four samples | 2.391 |
0.077 |
Nil |
| Citrobacter spp. | All the four samples | 3.448 |
0.022 |
Tukey’s HSD test was not significant |
| Proteus spp. | Pus and tissue | 0.717 |
0.403 |
Nil |
| Enterobacter spp. | All the four samples | 5.714 |
0.002 |
Blood-urine: 0.001 |
| Serratia spp. | Blood, pus and tissue | 5.096 |
0.01 |
Blood-pus: 0.016 |
| Pus-tissue: 0.031 | ||||
| *Please refer to the paragraph for the explanation of this table | ||||
Statistical analysis of the same microbes isolated from different samples: This analysis was performed on those microorganisms that were isolated from more than one sample to find out whether a microorganism behaves the same or differently for the same set of antibiotics in different samples. Microorganisms were analyzed, samples from which they were isolated and the results of statistical analysis are given in Table 17.
Table 17 shows the statistical analysis of microbes isolated from different samples. As shown in the table, it is clear that GPCs, i.e., S. aureus, MRSA and Enterococcus spp., behaved the same in all samples. They showed the same pattern of resistance irrespective of the sample type, although certain changes in the pattern were observed for some antibiotics individually which can be seen from the culture sensitivity tables of each sample, but collectively their means were the same. The changes for certain antibiotics will be discussed later in the statistical analysis of antibiotics section. Among GNRs, A. baumannii, E. coli and Proteus spp., showed the same resistance pattern in each sample, i.e., their resistance means were the same, although certain modifications for individual antibiotics are possible, which will also be discussed later.
While some GNRs were showing different resistance patterns in one or more samples. The significance value for P. aeruginosa is 0.001 (from Table 17), which reveals that it showed a different resistance pattern in any one of the samples. So from the post-hoc test (also shown in Table 17), it is clear that the P. aeruginosa isolated from blood samples was behaving differently than those isolated from pus and tissue samples. And by comparing their means from the descriptive table (not shown here), the P. aeruginosa from the blood sample generated a lower mean than those in the other samples. It is because it was showing fluctuations in resistance and sensitivity patterns for some antibiotics in the blood sample as compared to other samples, i.e., in a blood sample, it was 100% sensitive to some antibiotics, e.g., CIP, IMP, MEM and TZP, etc. and was resistant to them in pus and tissue samples and showed 50% resistance and 50% sensitivity for these antibiotics in urine sample. Therefore, the resistance means of P. aeruginosa in the blood sample was lower than those isolated from other samples.
Klebsiella pneumoniae (from Table 17) also showed differences in the resistance pattern isolated from any one of these samples, as its significance value is 0.014, which is smaller than 0.05, so, according to the alternative hypothesis, the means of the groups are not the same and there will be any one or more groups that showed variation. So from a post-hoc test, K. pneumoniae isolated from urine sample showed differences from those isolated from blood and tissue samples. The resistance mean of K. pneumoniae isolated from urine sample was small as compared to those isolated from other samples. It is because certain antibiotics such as DOX, TGC and PB were not used in antibiotic susceptibility testing of urine
samples, therefore, K. pneumoniae isolated from urine samples has a lower resistance mean than those isolated from other samples. Also, the K. pneumoniae isolated from a urine sample showed different resistance patterns for some antibiotics, just like the P. aeruginosa isolated from the blood samples. K. pneumoniae from the urine sample was sensitive to certain antibiotics, e.g., CIP, IMP, MEM, SCF and TZP, while it was resistant to these antibiotics in other samples.
The statistical analysis on Citrobacter spp. (from Table 17) is also significant because its sig. value is 0.022, which is smaller than 0.05, so there is one or more samples in which Citrobacter spp., produce different resistance mean. The post-hoc test, i.e., Tukey’s HSD, was not significant in this analysis, so LSD was used as an alternative. So, according to the LSD results and the descriptive table (both not shown here), it was concluded that Citrobacter spp., isolated from blood and urine samples behaved differently than those isolated from pus and tissue samples. This is because Citrobacter spp., isolated from blood and urine samples was sensitive to some antibiotics e.g., CIP (50%), IMP (50% for urine) and TZP (50% for urine) and was resistant to these antibiotics in pus and tissue samples.
Enterobacter spp., isolated from urine and blood samples also showed a difference in resistance pattern, as revealed from Table 17, because its significance value was 0.002 and the post-hoc test showed a difference in blood and urine samples. It is also because the Enterobacter spp., isolated from the blood sample was (100%) resistant to certain antibiotics, e.g., IMP, MEM, SCF and TZP, while sensitive to those isolated from other samples. And those isolated from urine samples were sensitive to CIP, while those isolated from other samples were resistant to CIP. Therefore, there is a major difference between the means of Enterobacter spp. (from the descriptive table, not shown here) isolated from blood samples and those isolated from other samples. Note that the test of Homogeneity of variance was violated in this case, so the results of Welch’s test were used as an alternative, which was also significant.
In the case of Serratia spp., the significance value of the ANOVA results is 0.010 (from Table 17), which is smaller than 0.05, so by accepting the alternative hypothesis, there is a group that is different than others and that was Serratia spp., isolated from pus samples as identified by post-hoc test (also from Table 17). So it means that Serratia spp., isolated from pus samples were behaving differently than those isolated from other samples. Serratia spp., isolated from pus samples was shown to be 100% resistant to those antibiotics that were slightly resistant or even sensitive to other GNRs or Serratia spp., isolated from other samples. Some of those antibiotics, as an example, are IMP, MEM, SCF and TZP. This may be either because only 2 samples of Serratia spp., were isolated from pus and if these were both resistant they would give 100% resistance, or because the Serratia spp., causing pus infections are more susceptible to antimicrobial resistance than those causing blood or tissue infections.
Statistical analysis of antibiotics used against many groups of microorganisms: Here comes the idea of a broad and narrow spectrum of antibiotics. Many of the antibiotics used in this study were broad-spectrum. Some of them were used as broad-spectrum antibiotics for GPCs only, some for GNRs only and some broad-spectrum antibiotics were used for both GPCs and GNRs. The aim of this analysis was to find the difference between groups that show a different pattern of resistance than other groups for which the same antibiotics are used. Table 18 shows the list of antibiotics analyzed, the list of microbes for which each antibiotic was used and the results of the statistical analysis.
Table 18 shows the broad-spectrum antibiotics used, on which statistical analysis was performed to find out whether the antibiotics used for a wide range of microorganisms are resistant to all of them or whether there are some microbes that are still sensitive to them. Not all antibiotics used in this study were included in this analysis because some of them were narrow-spectrum or some were intrinsically resistant to many groups. And two antibiotics, i.e., TGC and PB were sensitive to all microorganisms for which they were used (except Proteus spp., Providencia spp. and Serratia spp.). Only those antibiotics that were broad-spectrum and showed fluctuations in their resistance pattern were analyzed.
| Table 18: | Statistical analysis of antibiotics used for different groups of microorganisms | |||
ANOVA results |
||||
| Antibiotics | Microorganisms | F |
Significance |
post-hoc results (Tukey’s HSD) |
| Only for GPCs | ||||
| ERY | All the three GPCs | 13.855 |
0.009 |
S. aureus-MRSA: 0.040 |
| S. aureus-Enterococcus spp.: 0.009 | ||||
| LNZ | All the three GPCs | 0.525 |
0.617 |
Nil |
| P | S. aureus and Enterococcus spp. | 11.014 |
0.029 |
Nil |
| VAN | All the three GPCs | 1 |
0.422 |
Nil |
| For both GPCs and GNRs | ||||
| COT | S. aureus, MRSA, E. coli, A. baumannii, K. pneumoniae, Proteus spp., Citrobacter spp., Enterobacter spp. and Serratia spp. |
1.503 |
0.215 |
Nil |
| DOX | All three GPCs and E. coli, | 3.758 |
0.008 |
S. aureus-Serratia spp.: 0.045 |
| K. pneumoniae, A. baumannii, | MRSA-Serratia spp.: 0.014 | |||
| Proteus spp., Citrobacter spp., | A. baumannii-Serratia spp.: 0.028 | |||
| Enterobacter spp. and Serratia spp., | ||||
| among GNRs | ||||
| GEN | S. aureus, MRSA, P. aeruginosa, | 1.374 |
0.257 |
Nil |
| E. coli, A. baumannii, K. pneumoniae, | ||||
| Proteus spp., Citrobacter spp., | ||||
| Enterobacter spp. and Serratia spp., | ||||
| CIP | All from above (GEN) | 0.658 |
0.738 |
Nil |
| Only for GNRs | ||||
| AK | P. aeruginosa, E. coli, A. baumannii | 3.034 |
0.024 |
Tukey’s HSD was not significant |
| K. pneumoniae, Proteus spp., | ||||
| Citrobacter spp., Enterobacter spp. | ||||
| and Serratia spp. | ||||
| CRO | Same for AK except P. aeruginosa | 1.817 |
0.155 |
Nil |
| FEP | Same for AK | 1.339 |
0.284 |
Nil |
| IMP | Same for AK | 0.947 |
0.494 |
Nil |
| MEM | Same for AK | 1.606 |
0.191 |
Nil |
| SCF | Same for AK except P. aeruginosa | 2.336 |
0.079 |
Nil |
| TZP | Same for AK | 1.089 |
0.407 |
Nil |
| CAP | Same for AK except P. aeruginosa and A. baumannii |
0.849 |
0.536 |
Nil |
| *AK: Amikacin, FEP: Cefepime, CRO: Ceftriaxone, CAP: Chloramphenicol, CIP: Ciprofloxacin, COT: Co-trimoxazole, DOX: Doxycycline, ERY: Erythromycin, GEN: Gentamicin, IMP: Imipenem, LNZ: Linezolid, MEM: Meropenem, P: Penicillin, SCF: Sulbactum-Cefoperazone, TZP: Tazobactum-Peperacillin and VAN: Vancomycin | ||||
As from Table 18, it is clear that antibiotics that were used only for GPCs, ERY showed differences in resistance patterns. Its significance value from ANOVA results was 0.009, so according to an alternative hypothesis, there must be a group that is different from others. So from the post-hoc test, it is S. aureus that gives different resistance means to ERY than MRSA and Enterococcus spp. It is because MRSA and Enterococcus spp., were found to be highly resistant to ERY, while S. aureus was slightly resistant to it. As, P also showed a difference, i.e., its significance value is 0.029. It is intrinsically resistant to MRSA, therefore, only S. aureus and Enterococcus spp., were used in the analysis. Its significance value reveals that S. aureus and Enterococcus spp., didn’t give the same pattern of resistance (post-hoc test was not applied because only two groups were analyzed). So, by looking at their means from the descriptive table (not shown here) and culture sensitivity table, it is concluded that S. aureus was highly resistant to P as compared to Enterococcus spp. Other antibiotics that were used only for GPCs behaved the same, i.e., LNZ and VAN were found sensitive to all GPCs for which they were used in this study, which can also be confirmed by looking through the culture sensitivity tables of each sample. Note that the homogeneity of variance was violated for analyzing LNZ and VAN, but Welch’s test used as an alternative was also not significant for LNZ and didn’t give any result for VAN because any one group’s mean was 0 (means 0% resistant).
Among the antibiotics that were used for both GPCs and GNRs (from Table 18), only DOX showed significant results. Its significance value is 0.005, which means that there is a difference between one or more groups. From the post-hoc test, Serratia spp., showed differences with S. aureus, MRSA and A. baumannii. This is because of the major difference between their means because among GPCs, i.e., S. aureus and MRSA and among GNRs, A. baumannii was found sensitive for DOX while other GNRs and Enterococcus spp., were slightly resistant and Serratia spp., was highly resistant to DOX. Therefore, there was a major difference between their means. Note that homogeneity of variance was violated in this case, so, the result of Welch’s test was used as an alternative, which was also significant. Other antibiotics used for both GPCs and GNRs showed the same pattern of resistance as a whole, but by looking at their pattern individually for each microbe, there are some fluctuations, but these are very minor changes and in ANOVA, their means are compared therefore, it showed no difference. The COT showed slight sensitivity to GPCs i.e., S. aureus and MRSA, while being slightly resistant to some GNRs and highly resistant to some GNRs. And CIP showed slight resistance for GPCs and high resistance for GNRs present in this study.
The GEN showed no difference according to ANOVA results because its significance value is 0.254 (from Table 18), but homogeneity of variance was violated in this case and Welch’s test, used as an alternative, showed the significant results. Therefore, different post-hoc tests were applied to identify the error. Tukey’s HSD and Dunnett’s test also didn’t give any significant results, but the LSD test showed a difference for S. aureus with E. coli, K. pneumoniae and Citrobacter spp. The means from the descriptive table (not shown here) and culture sensitivity tables for each sample conclude that GEN was slightly sensitive for S. aureus and MRSA and slightly resistant for A. baumannii, Proteus spp., P. aeruginosa, Serratia spp. and Enterobacter spp., while it was highly resistant to E. coli, K. pneumoniae and Citrobacter spp. This difference between their means was very minor, therefore ANOVA, Tukey’s test and Dunnett’s test didn’t give statistically significant results.
Among the broad-spectrum antibiotics that were used only for GNRs (Table 18), only AK showed a difference in the resistance pattern of the microbes against which it was used. Its significance value from ANOVA results is 0.024, which means that any one or more groups are showing a difference in their susceptibility. The post-hoc test used (Tukey’s test) didn’t show any significant results, but another post-hoc test, i.e., the LSD test, was also performed as an alternative. The LSD test showed differences in E. coli and Enterobacter spp., with other GNRs. The AK was showing many fluctuations in its resistance pattern for each microbe within each sample. So collectively, by comparing the means of microbes used in analysis and for which AK was used, it was concluded that AK was found to be slightly sensitive to E. coli, Enterobacter spp. and Proteus spp., while slightly resistant to P. aeruginosa, A. baumannii, K. pneumoniae, Serratia spp. and Citrobacter spp. (which was 50% resistant and 50% sensitive according to its mean).
Other antibiotics that were used only for GNRs (Table 18), behaved the same for all microbes for which they were used in this study. Although there are some changes in their pattern when looking at each microbe in each sample individually, collectively, their means are the same for the microbes used, so they didn’t give any significant results in the ANOVA. The susceptibility pattern of these antibiotics according to their mean values from the descriptive table (not shown here) are as follows: CRO was found resistant to all GNRs for which it was used except for Proteus spp.,, for which it was found sensitive. The FEP was found to be resistant to all GNRs, but it was slightly sensitive to P. aeruginosa isolated from blood and urine samples, as evident from the ANOVA results of P. aeruginosa in the statistical analysis on microbes section (Table 17). The IMP was found to be sensitive to P. aeruginosa, K. pneumoniae, Enterobacter spp., E. coli, Citrobacter spp. and Serratia spp. and highly sensitive to Proteus spp., while resistant to A. baumannii. The MEM was found sensitive for P. aeruginosa, E. coli, Proteus spp. and Enterobacter spp., while 50% resistant and 50% sensitive to Serratia spp. and resistant to A. baumannii, K. pneumoniae and Citrobacter spp. The SCF was found to be sensitive to E. coli, Proteus spp. (highly sensitive), Citrobacter spp.
and Enterobacter spp., while resistant to A. baumannii, K. pneumoniae and Serratia spp. The TZP was found to be sensitive to P. aeruginosa, E. coli, Proteus spp. and Enterobacter spp., while resistant to A. baumannii, K. pneumoniae, Citrobacter spp. and Serratia spp. The CAP was found to be sensitive to all microbes for which it was used except for Proteus spp. Note that the homogeneity of variance was violated for the analysis of CAP and the results of Welch’s test were significant, but the post-hoc tests (Tukey’s HSD and LSD) were also not significant, therefore it was considered non-significant as analyzed by ANOVA.
DISCUSSION
The current study was conducted on different clinical isolates from different sites for prevalence and antibiotic susceptibility pattern. A total of 2209 clinical samples were collected and studied in the Tertiary Care Hospital, Peshawar, in which 43.8% were blood samples, 14.9% were pus samples, 9.8% were tissue samples and 31.4% were urine samples. Among these, only 21.7%, i.e., 104 samples, produced bacterial colonies, 56 of these culture-positive samples produced the growth of two microbes, (i.e., polymicrobial infection), so the total number of microbes in this study was 535. By looking through the age and gender wise distribution of these culture positive samples, it was concluded that microbial infections can hit anyone at any age, irrespective of age and gender.
Of the total 968 samples of blood, only 83 were culture positive due to sterile nature of blood and selective media used during the study. These results are in line with the study conducted by Rajeevan et al.22, in which only 113 samples came out as culture positive from 1196 total samples. As 18, out of 83 isolates from blood were GPCs, i.e., S. aureus and MRSA and the rest, i.e., 65, were GNRs. The GPCs were highly resistant to penicillin, followed by erythromycin and co-trimoxazole and highly sensitive to linezolid and vancomycin. S. aureus was also found to be sensitive to cloxacillin and co-amoxiclav while MRSA was intrinsically resistant to them. Among GNRs, the most prevalent was S. typhi, followed by K. pneumoniae, A. baumannii, P. aeruginosa, Citrobacter spp., E. coli, Serratia spp. and Enterobacter spp. Salmonella typhi, was found sensitive to antibiotics used in the study except, ampicillin and ciprofloxacin followed by ceftriaxone and co-trimoxazole. Pseudomonas aeruginosa isolated from blood samples was found to be highly resistant to ceftazidime and highly sensitive to tazobactam-piperacillin, followed by others. Other GNRs isolated were found to be highly resistant to azithromycin, followed by ceftriaxone, ciprofloxacin and meropenem and highly sensitive to tigecycline and polymyxin-B. The study was conducted by Abebaw et al.23, at the University of Gondar from March to May, 2013. Out of the confirmed cases, the most commonly isolated microbes were more Gram-positive than Gram-negatives. Gram-positives were found highly resistant to ampicillin and erythromycin and highly sensitive to vancomycin and ciprofloxacin, while Gram-negatives were found highly resistant to ampicillin and amoxicillin and highly sensitive to norfloxacin and ciprofloxacin. As this was 2013 data, most of the antibiotics that were sensitive at that time are now highly resistant in this study, such as Ciprofloxacin.
The high number of pus and tissue samples in this study indicates that SSTIs are a major cause of hospitalization in both developed and underdeveloped countries and can cause tissue damage. This can be proved by a study conducted in a developed country (i.e., US) in which 2.4 million patients were identified to be infected with skin and soft tissue infections in the year 2000 and a total of 3.3 million patients experienced SSTIs in 2012. A study conducted in an underdeveloped country (i.e., Pakistan) identified 72 patients experiencing necrotizing soft tissue infections in 2021 in a tertiary care hospital24,25. A total of 330 samples of pus and 217 samples of tissue were collected, of which 216 pus samples and 104 tissue samples were culture-positive. In some cases, they both produced polymicrobial colonies, i.e., 27 samples of pus and 29 samples of tissue, so the total number of microbial isolates was 243 in the pus samples and 133 in the tissue samples. The microbial isolates and their antibiotic susceptibility pattern were almost the same in both samples. The most highly identified GPCs in both cases were S. aureus (most of them were MRSA), followed by Enterococcus spp. Among GNRs, the most highly identified microorganism was P. aeruginosa, followed by A. baumannii, K. pneumoniae, E. coli and Proteus spp. The GPCs identified in both of these samples gave the following antibiotic susceptibility pattern: Staphylococcus aureus and MRSA were highly resistant to penicillin, ciprofloxacin and erythromycin, while sensitive to vancomycin, linezolid, rifampicin and doxycycline. S. aureus was also found to be sensitive to cloxacillin and co-amoxiclav. Enterococcus spp., was found to be highly resistant to erythromycin, followed by penicillin and doxycycline, while sensitive to linezolid and vancomycin. Among GNRs: Pseudomonas aeruginosa was highly resistant to ceftazidime, followed by ciprofloxacin, meropenem, tobramycin, gentamycin and cefepime (their order is change for both), while sensitive to polymyxin-B and tazobactam-piperalin (slightly resistant in the pus sample). Other GNRs were highly resistant to ciprofloxacin, ceftriaxone, cefepime and Tazobactam-piperacillin, while sensitive to polymyxin-B and tigecycline (except for those for which they were intrinsically resistant) and chloramphenicol. A study conducted by Waheed et al., had the same results. Among the cultured positive samples collected from the site of SSTIs, the organisms identified were gram-positives in which S. aureus was dominant and gram-negatives in which E. coli was dominant. Gram-positives were found highly sensitive to vancomycin, fusidic acid, linezolid, amikacin, chloramphenicol, gentamicin and tigecycline. Gram-negatives were highly resistant to cephalosporin, ampicillin, erythromycin and co-trimoxazole and slightly resistant to ticarcillin, tazobactam-piperacillin, amikacin and gentamicin26. Proteus spp., were only isolated from UTIs, but in this study they were isolated from pus and tissue samples, which means that they can now cause SSTIs or act as a secondary pathogen in an already infected site or wound. This Proteus spp., is also found to be resistant to some antibiotics, but it is sensitive to many antibiotics, so timely prevention and treatment can stop the emergence of multidrug resistant Proteus spp.
Out of a total of 694 urine samples collected, 76 were culture-positive. As compared to other samples in this study, the culture-positive urine samples were from both the male and female populations. The isolated microbes were identified as Enterococcus spp., among GPCs and E. coli, followed by K. pneumoniae, P. aeruginosa, Enterobacter spp. and Citrobacter spp., among GNRs. The GPCs were highly resistant to levofloxacin, fosfomycin and ciprofloxacin, while sensitive to linezolid and vancomycin, followed by nitrofurantoin, penicillin and erythromycin. Pseudomonas aeruginosa among GNRs was resistant to ceftazidime and sensitive to polymyxin-B, followed by others. Other GNRs were resistant to ciprofloxacin, ceftriaxone and cefepime, while sensitive to imipenem, amikacin and meropenem. A similiar study conducted by Calzada et al.27, collected urine samples from 113 patients at Saint Joseph Kingdom Hospital, Kitgum, Uganda. From these samples, 100 microorganisms were identified, of which the most highly isolated microbes were Enterococcus spp., among gram-positive bacteria and E. coli among gram-negative bacteria. Gram-positive were found highly (100%) resistant to penicillin, cefoxitin, kanamycin, clindamycin, erythromycin and trimethoprim and sensitive to linezolid, vancomycin and teicoplanin. Gram-negatives were found to be resistant to trimethoprim/sulfamethoxazole, ampicillin, piperacillin, cefalexin, cefepime and ceftazidime and sensitive to tigecycline, ceftriaxone, fosfomycin and nitrofurantoin.
The statistical analysis, i.e., One-way ANOVA along with the homogeneity of variance test, Welch’s test and post-hoc tests performed in this study through different dimensions, produced various results. The analysis conducted on the GPCs and GNRs of each sample gave the following results: The set of antibiotics used for both GPCs (S. aureus and MRSA) and all GNRs (except S. typhi and P. aeruginosa) isolated from blood samples follow the same pattern of resistance, i.e., there means show no difference as a whole. The GPCs isolated from pus and tissue samples also follow the same pattern, but their GNRs broke out the trend. The GNRs of the pus sample gave significant result with a group that was giving a different pattern from others and that group was identified as Serratia spp., from the post-hoc test. The GNRs of tissue samples also gave significant results, but any one group could not be identified because they were all behaving differently from each other. The analysis of the GNRs of urine samples didn’t give any significant results, which means that the means of resistance were the same for the same antibiotics used. This concluded that the GPCs (S. aureus and MRSA) of all three samples were giving the same pattern as the GNRs of blood and urine samples, while there were groups in the GNRs of pus and tissue samples that were behaving differently than others.
A statistical analysis was also performed on each microbe that was isolated from more than one sample to study its behavior with the same set of antibiotics in different samples. This analysis gave the following results: The GPCs, i.e., S. aureus, MRSA and Enterococcus spp., behaved the same in all samples from which they were isolated. While in GNRs A. baumannii, E. coli and Proteus spp., produced the same means of resistance, P. aeruginosa, K. pneumoniae, Citrobacter spp., Enterobacter spp. and Serratia spp., produced different patterns in any one sample from which they were isolated. P. aeruginosa isolated from blood samples, K. pneumoniae isolated from urine samples, Citrobacter spp. and Enterobacter spp., isolated from blood and urine samples and Serratia spp. isolated from pus samples were giving a different resistance pattern from those isolated from other samples. It means that the same microbe can behave differently for the same antibiotic at different sites of infection. So before beginning any treatment, the site of infection should be kept in mind and only those antibiotics should be preferred that showed high susceptibility in in-vitro reports.
Another analysis was performed on antibiotics that are broad-spectrum and are used for different groups of microorganisms or genera of bacteria. Broad-spectrum antibiotics act on a target that is present in many microbes, so they are effective for those microbes that contain the same target (although the concept of natural resistance also exists). But the emergence of antimicrobial-resistant microbes or multidrug-resistant bugs (superbugs) and the rapid transfer of these resistant genes to the same species genetically and to different species through horizontal gene transfer (HGT) are reducing the efficacy of these drugs. The aim of this analysis was to check the resistance pattern of antibiotics for which they are used, i.e., an antibiotic identified as resistant is either resistant to all those microbes for which it is used as a broad-spectrum antibiotic or there are some microbes that are still sensitive to it and vice versa.
The statistical analysis performed on broad-spectrum antibiotics showed the following results: Among the antibiotics that were used and analyzed only for GPCs, linezolid and vancomycin showed the same pattern and they both were sensitive for the GPCs isolated in this study. Erythromycin and penicillin showed significant results. Erythromycin was found to be highly resistant to MRSA and Enterococcus spp., while it was slightly resistant to S. aureus and penicillin was found resistant to S. aureus, while it was sensitive to Enterococcus spp., so it is concluded that penicillin can be used to treat Enterococcus-causing infections. Among antibiotics that were used for both GPCs and GNRs, doxycycline produced significant ANOVA results because it was found sensitive to S. aureus and MRSA among GPCs and A. baumannii among GNRs, while it was slightly and highly resistant to other GNRs. Among the other antibiotics in this category, gentamicin also produced a significant post-hoc LSD test and it was also found to be slightly sensitive to S. aureus and MRSA and slightly and highly resistant to GNRs, while co-trimoxazole and ciprofloxacin showed the same pattern or the same mean in ANOVA and they were found resistant for all microbes for which they were used in this study. Among the broad-spectrum antibiotics that were used only for GNRs, only AK produced significant results because it was found to be slightly sensitive for some GNRs and slightly resistant for others. Other antibiotics such as ceftriaxone, cefepime, imipenem, meropenem, sulbactam-cefoperazone, tazobactam-piperacillin and chloramphenicol were found to produce the same means in ANOVA results. Among them, ceftriaxone was found resistant to all and cefepime as well (except P. aeruginosa) and chloramphenicol was found sensitive to all (except Proteus spp. and A. baumannii), while others, i.e., imipenem, meropenem, sulbactam-cefoperazone and tazobactam-piperacillin, were sensitive to some microbes and resistant to some microbes. So they should not be used against the microbes to which they were found resistant.
The difference in the resistance pattern of the same microbe from different samples for the same antibiotic is due to the spread of resistant genes in the community, the individual’s immunity and environment, the community’s behavior towards antibiotics, the contaminated environment and a lack of personal care. These are some factors that generate multidrug resistant bugs in the environment. It is not only limited to their production, but these genes can also be transferred from one species to another through the environment or by using living organisms as a carrier or vector. As confirmed by this study, the broad-spectrum antibiotics that were first effective for microbes are now either resistant to all of them or still slightly sensitive to some of them and it will take no time for them to become slightly resistant and then highly resistant from being slightly sensitive. Some of the microbes isolated from this study were multidrug resistant, i.e., MRSA, E. coli, P. aeruginosa, K. pneumoniae and A. baumannii. The high amount of E. coli in urine samples indicates its multidrug resistance nature. Serratia spp., is also an emerging multidrug-resistant bug, as confirmed by this study as well.
The results obtained in this study might contain some minor errors due to the following reasons, they may be due to any contamination during sample collection and laboratory processing and certain parameters were not available during the study, which could cause changes in the resistance pattern. This study was conducted at a Tertiary Care Hospital, Peshawar, so the samples were taken not only from Peshawar, but from other cities in the province of Khyber Pakhtunkhwa as well, so the results and findings of this study are applicable all over the province of Khyber Pakhtunkhwa. From a future perspective, different studies could also be conducted based on the results of this study, such as molecular characterization and PCR analysis of multidrug-resistant microbes. A study could also be conducted on the resistance patterns of other antibiotics that are used in practical life but were not used in this study or on other microbial pathogens that were not isolated in this study.
CONCLUSION
As a developing country, Pakistan is prone to many infectious and chronic diseases. Among the infectious diseases used in this study were bloodstream infections, skin and soft tissue infections and urinary tract infections. All of these infections can cause both hospital- and community-acquired infections. These all range from mild to severe and life-threatening and this is because of their multidrug-resistant infectious agents. In the present study, blood and urine samples produced a lower number of culture-sensitive samples as compared to their total number of samples than pus and tissue samples, so SSTIs are the major cause of hospitalization. The microbes identified confirmed the presence of some new pathogens that are not only causing infections at different sites, i.e., other than the typical site of infection but are also resistant to some antibiotics. The antibiotic susceptibility testing and statistical analysis on isolated microbes concluded that most of them were resistant to most of the antibiotics used. So the antibiotics that are causing high resistance patterns, such as ciprofloxacin, erythromycin, ampicillin, ceftazidime and ceftriaxone should be stopped and new antibiotics should be used. It is also concluded that a microbial examination is necessary before giving or using any antimicrobial therapy because the same microbes behave differently in different sites of infection and antibiotics behave differently for different microbes in different sites as well. Precautionary measures should be taken according to the risk factors for different kinds of infections to prevent them and also to stop the production and spread of multidrug-resistant bugs.
SIGNIFICANCE STATEMENT
Multidrug resistance is a new disaster in the healthcare setting that originated in the past and is still evolving and getting worse day by day especially in developing countries. This study was thus conducted on the same issue to track new patterns of antibiotic resistance. Along with the resistance pattern, this study also discovered the secondary pathogenic character (a character to cause an infection in an already
infected site by another pathogen) of certain microbes that were known to cause infection only at a certain site. The results of this study are important for a country's timely prevention of multi-drug-resistant and secondary pathogenic microbes. The analysis in this study will provide a new way for statistical analysis of resistance patterns.
ACKNOWLEDGMENTS
We admire the help and guidance of our supervisor, Dr. Nadia Bibi, Assistant Professor at the Department of Microbiology. Special thanks to Maj. Anees Ur Rehman for his remarkable consideration to make it possible and easy to work in the microbiology laboratory in Tertiary Care Hospital, Peshawar. We are also thankful to other hospital staff, especially Sep. Ali Raza, Frontier Corps. Khalid Hussain and Dr. Shehnaz, for their help during experimentation.
REFERENCES
- Gohel, K., A. Jojera, S. Soni, S. Gang, R. Sabnis and M. Desai, 2014. Bacteriological profile and drug resistance patterns of blood culture isolates in a tertiary care nephrourology teaching institute. BioMed Res. Int., 2014: 153747.
- Parajuli, N.P., H. Parajuli, R. Pandit, J. Shakya and P.R. Khanal, 2017. Evaluating the trends of bloodstream infections among pediatric and adult patients at a teaching hospital of Kathmandu, Nepal: Role of drug resistant pathogens. Can. J. Infect. Dis. Med. Microbiol., 2017: 8763135.
- Deku, J.G., M.P. Dakorah, S.Y. Lokpo, V.N. Orish and F.A. Ussher et al., 2019. The epidemiology of bloodstream infections and antimicrobial susceptibility patterns: A nine-year retrospective study at St. Dominic Hospital, Akwatia, Ghana. J. Trop. Med., 2019: 6750864.
- Kolesnichenko, S.I., A.V. Lavrinenko and L.L. Akhmaltdinova, 2021. Bloodstream infection etiology among children and adults. Int. J. Microbiol., 2021: 6657134.
- Vasudeva, N., P.S. Nirwan and P. Shrivastava, 2016. Bloodstream infections and antimicrobial sensitivity patterns in a tertiary care hospital of India. Ther. Adv. Infect., 3: 119-127.
- Al-Asady, F.M., D.A. Al-Saray and A.W. Obed, 2020. Incidence of septicemia. Etiology and antimicrobial susceptibility testing among patients admitted to tertiary care hospital. J. Infect. Dev. Countries, 14: 1387-1394.
- Kaye, K.S., L.A. Petty, A.F. Shorr and M.D. Zilberberg, 2019. Current epidemiology, etiology, and burden of acute skin infections in the United States. Clin. Infect. Dis., 68: S193-S199.
- Ramakrishnan, K., R.C. Salinas and N.I.A. Higuita, 2015. Skin and soft tissue infections. Am. Fam. Physician, 92: 474-483.
- Miller, L.G., D.F. Eisenberg, H. Liu, C.L. Chang and Y. Wang et al., 2015. Incidence of skin and soft tissue infections in ambulatory and inpatient settings, 2005-2010. BMC Infect. Dis., 15: 362.
- Esposito, S., S. Noviello, F. de Caro and G. Boccia, 2018. New insights into classification, epidemiology and microbiology of SSTIs, including diabetic foot infections. Le Infezioni Med., 26: 3-14.
- Agrawal, S.K., A. Panigrahy, S. Perumalla, A. Kapil and B. Dhawan, 2018. Microbiological profile and antibiotic resistance pattern of skin and soft-tissue infections: A study from Northern India. J. Lab. Physicians, 10: 471-472.
- Montravers, P., A. Snauwaert and C. Welsch, 2016. Current guidelines and recommendations for the management of skin and soft tissue infections. Curr. Opin. Infect. Dis., 29: 131-138.
- Khan, I., N. Sarwar, B. Ahmad, S. Azam and N. Rehman, 2017. Identification and antimicrobial susceptibility profile of bacterial pathogens isolated from wound infections in a teaching hospital, Peshawar, Pakistan. Adv. Life Sci., 5: 8-12.
- Foxman, B., 2002. Epidemiology of urinary tract infections: Incidence, morbidity and economic costs. Am. J. Med., 113: 5-13.
- Flores-Mireles, A.L., J.N. Walker, M. Caparon and S.J. Hultgren, 2015. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol., 13: 269-284.
- Kibret, M. and B. Abera, 2014. Prevalence and antibiogram of bacterial isolates from urinary tract infections at Dessie Health Research Laboratory, Ethiopia. Asian Pac. J. Trop. Biomed., 4: 164-168.
- Namdari, S., A. Farhadi, A. Khademalhoseini, A. Behzad-Behbahani and A. Moaddeb, 2021. Emergence of highly multidrug-resistant bacteria isolated from patients with infections admitted to public hospitals in Southwest Iran. Interdiscip. Perspect. Infect. Dis., 2021: 5265379.
- Tenover, F.C., 2006. Mechanisms of antimicrobial resistance in bacteria. Am. J. Med., 119: S3-S10.
- Nikaido, H., 2009. Multidrug resistance in bacteria. Annu. Rev. Biochem., 78: 119-146.
- Alemu, A., F. Moges, Y. Shiferaw, K. Tafess, A. Kassu, B. Anagaw and A. Agegn, 2012. Bacterial profile and drug susceptibility pattern of urinary tract infection in pregnant women at University of Gondar Teaching Hospital, Northwest Ethiopia. BMC Res. Notes, 5, No. 1.
- Idrees, F., K. Jabeen, M.S. Khan and A. Zafar, 2009. Antimicrobial resistance profile of methicillin resistant staphylococcal aureus from skin and soft tissue isolates. J. Pak. Med. Assoc., 59: 266-269.
- Rajeevan, S., S.M. Ahmad and P.T. Jasmin, 2014. Study of prevalence and antimicrobial susceptibility pattern in blood isolates from a tertiary care hospital in North Kerala, India. Int. J. Curr. Microbiol. Appl. Sci., 3: 655-662.
- Abebaw, A., H. Tesera, T. Belachew and G.D. Mihiretie, 2018. The bacterial profile and antibiotic susceptibility pattern among patients with suspected bloodstream infections, Gondar, North-West Ethiopia. Pathol. Lab. Med. Int., 10: 1-7.
- Tun, K., J.F. Shurko, L. Ryan and G.C. Lee, 2018. Age-based health and economic burden of skin and soft tissue infections in the United States, 2000 and 2012. PLoS ONE, 13.
- Majid, H.J., M.I. Anwar, M.Z. Khalid, Shafique-Ur-Rehman and M.A. Jameel, 2021. Predictors of mortality in necrotizing soft tissue infections in adults-experience at a tertiary care hospital in Pakistan. Proceedings, 35: 24-30.
- Waheed, A., A. Amar, I. Afridi, S. Rauf and Mehran, 2020. Bacteriological profile and antibiotics susceptibility patterns of complicated skin and skin structure infections in tertiary care hospitals, Peshawar. J. Pak. Assoc. Dermatol., 30: 580-586.
- Calzada, F.C., J.J. Aguilera-Correa, J.C. González, J.E. Moreno, D.R. Biosca and R. Pérez-Tanoira, 2022. Urinary tract infection and antimicrobial susceptibility of bacterial isolates in Saint Joseph Kitgum Hospital, Kitgum, Uganda. Antibiotics, 11: 504.
How to Cite this paper?
APA-7 Style
Naveed,
M., Riaz,
A., ,
M., Bibi,
N. (2023). Prevalence and Antimicrobial Susceptibility Testing and Analysis of Microbes Isolated from Different Clinical Samples. Asian Journal of Biological Sciences, 16(3), 372-400. https://doi.org/10.3923/ajbs.2023.372.400
ACS Style
Naveed,
M.; Riaz,
A.; ,
M.; Bibi,
N. Prevalence and Antimicrobial Susceptibility Testing and Analysis of Microbes Isolated from Different Clinical Samples. Asian J. Biol. Sci 2023, 16, 372-400. https://doi.org/10.3923/ajbs.2023.372.400
AMA Style
Naveed
M, Riaz
A,
M, Bibi
N. Prevalence and Antimicrobial Susceptibility Testing and Analysis of Microbes Isolated from Different Clinical Samples. Asian Journal of Biological Sciences. 2023; 16(3): 372-400. https://doi.org/10.3923/ajbs.2023.372.400
Chicago/Turabian Style
Naveed, Momina, Aruba Riaz, Mahnoor , and Nadia Bibi.
2023. "Prevalence and Antimicrobial Susceptibility Testing and Analysis of Microbes Isolated from Different Clinical Samples" Asian Journal of Biological Sciences 16, no. 3: 372-400. https://doi.org/10.3923/ajbs.2023.372.400

This work is licensed under a Creative Commons Attribution 4.0 International License.



