Macroscopic and Molecular Approaches for Identification of Fungi Associated with Azanza garckeana
| Received 12 Mar, 2024 |
Accepted 17 Apr, 2024 |
Published 30 Sep, 2024 |
Background and Objective: Azanza garckeana, is an important African shrub for its medical and economic benefits. However, more research is needed on the molecular identification of fungi associated with it. In this research, macroscopic morphology and molecular techniques were used to identify the genus of the fungal pathogens causing deterioration of goron tula fruit. Materials and Methods: Macroscopic identification was based on the characteristics of the fungus grown on the Potato Dextrose Agar plate and the molecular identification was based on the DNA of the fungal isolate. The DNA of the fungal isolate, GR-3E was characterized using Internal Transcribed Spacer 1 and 4 molecular markers and aligned by the Basic Local Alignment Search Tool for Nucleotide (BLASTN) 2.8.0 of National Centre for Biotechnology Information (NCBI) database. Based on sequence similarity, it was observed that isolate GR-3E was 98.96% identical to Aspergillus candidus. Results: The results indicated that the GR-3E isolate sequence was 100% identical to Aspergillus candidus isolate. These findings showed that Aspergillus candidus is one of the causal fungal pathogens of post-harvest rot infecting goron tula fruits. To determine the link between the isolates collected from this study, a phylogenetic tree was built. Conclusion: Some information on the fungus that causes goron tula decay has been provided by this study. This outcome is expected to give insights into a disease control strategy to mitigate post-harvest losses of goron tula fruits brought on by A. candidus and lay the groundwork for additional research on the potential risks associated with consuming infected goron tula.
| Copyright © 2024 Ikechi-Nwogu and Egere. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
There has been an increasing need to curb health issues such as menstruation abnormalities, sexually transmitted infections, high blood pressure, ulcers and infertility. The name goron tula (Azanza garckeana) is considered a miracle fruit that can be used to curb some of these health issues. This study explored the possibilities of identifying fungi pathogens associated with the goron tula (Azanza garckeana) fruits using basic molecular techniques, owing to the fact that plants are subject to diseases.
Azanza garckeana (F. Hoffm.) Exell and Hillc, referred to in Hausa as goron tula, meaning Kola of Tula, is a member of the Malvaceae family1. Only the Tula people in Nigeria's Kaltungo Local Government Area of Gombe State and Michika in Adamawa State have access to this tasty native tree species2. Azanza garckeana is a deciduous shrub with a stem diameter of up to 25 cm at breast height. Depending on the environment, the tree can reach heights of 3 to 15 m3. Mojeremane and Tshwenyane4 and Djarova et al.5 in their study, testified that the fruit has vitamin C, protein, carbohydrates and a few minerals including calcium, magnesium, potassium, manganese and iron. Yusuf et al.1 reported that it is a significant food and medicinal plant that is widely utilized to make herbal remedies in Northern Nigeria.
Azanza garckeana's greatest valuable resource is its fruits in terms of sweetness and medicinal value. Economically, goron tula are important fruits because they possess several health benefits. It is considered a miracle fruit that can be used to curb some health issues such as infertility6, low libido, vagina odour and lubrication. It has also reportedly been used in traditional medicine as an herbal remedy for diseases like cough, chest pains, irregular menstruation, sexually transmitted infections and hepatic impairments7.
This project aims to determine the fungi associated with the seeds of Azanza garckeana using molecular techniques.
MATERIALS AND METHODS
Study area: This research was carried out from 23rd October 2022 to 9th April 2023 at the University of Port Harcourt's Animal Science Molecular Laboratory in Choba, Rivers State, Nigeria.
Source of plant material: Twenty fresh samples of Azanza garckeana seeds with contaminations were chosen at random from Tula in the Kaltungo Local Government Area, Gombe State. The samples were collected into sample bags, labelled and immediately taken to the laboratory.
Isolation of fungi from Azanza garckeana seeds using the blotter method: Fungal pathogens associated with A. garckeana seeds were isolated using a standard blotter method modified by Ikechi-Nwogu et al.8. The Petri dishes were lined with 3 layers of sterile filter papers, which were soaked in distilled water. The kola nuts were surface sterilized in a beaker using 70% ethanol, plated and incubated for 7 days at a temperature of 25+2°C. The organism was isolated at the Animal Science Molecular Laboratory, Faculty of Agriculture and the DNA was extracted at the same laboratory in the University of Port Harcourt, Choba, Rivers State, Nigeria.
Purification and sequencing of the PCR products were carried out at the International Institute of Tropical Agriculture (IITA) Ibadan where the most common fungal isolate was coded (GR-3E).
Morphological and microscopic characterization and identification: The morphological identification of isolates GR-3E was conducted by visually observing the mycelium and comparing their colonies for their diameters, colors, colors of conidia, reverse colors, texture, zonation and sporulation with Snowdon9 pictorial guides. The isolates were later subjected to microscopic analysis for identification using a Celestron electron binocular microscope (Guangzhou, China) at X40. Molecular characterization using the Internal Transcribed Spacer (ITS) marker and identification.
The Quick-DNATM Fungal/Bacterial MiniPrep Kit (Zymo Research Group, California, USA) was modified at the Regional Centre for Biotechnology and Bioresources (RCBB), University of Port Harcourt, Rivers State, Nigeria, in accordance with the manufacturer's instructions to extract the isolate GR-3E's genomic DNA. Thermo Fisher Scientific Inc., Wilmington, Delaware, USA, provided the Nanodrop 2000c spectrophotometer, which was used to measure the quantity and concentration of DNA from the GR-3E isolate. The ratio of absorbance at 280 nm to that at 260 nm was used to calculate the purity of the DNA. The modified Saghai-Maroof et al.10 approach was utilized to perform Agarose gel electrophoresis to further quantify the purity of the isolate GR-3E's DNA.
Statistical analysis: The sequence was analysed using the Molecular Evolutionary Genetics Analysis (MEGA) version 7.0.26 software and aligned using the Basic Local Alignment Search Tool for nucleotide (BLASTN) 2.8.0 version of the National Centre for Biotechnology Information (NCBI) Database.
RESULTS AND DISCUSSION
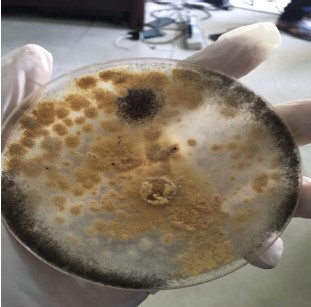
The results of the fungal isolation culture are presented in Fig. 1. One unidentified fungal organism GR-3E was isolated and found to be associated with Azanza garckeana. The isolates developed a yellow spore surrounded by a white spore that was visible to the eyes. From the photomicrograph, the isolate was identified as an Aspergillus spp. This slow-growing organism was white at first but later turned yellow. It has a circular form and sometimes it is punctiform, the elevation is flat and the margin is described as entire. The surface of this organism is dry and powdery. The conidia are white-yellow in colour.
DNA extraction and concentration determination: The genomic DNA of the isolates GR-3E of goron tula was successfully extracted. The NanoDrop result showed that the concentrations of the DNA of the isolates were 238.5 ng/μL. While the absorption peak of the 260/280nm readings was 1.86 and the 260/230nm readings were 1.52.
Polymerase Chain Reaction (PCR) and gel electrophoresis: The result of the amplified DNA or PCR band of the isolate GR-3E is presented in Fig. 2. The amplified DNA showed a band on the gel when observed under UV light. From the result, the ladder used indicated that the GR-3E isolate sequence had over 600 base pairs.
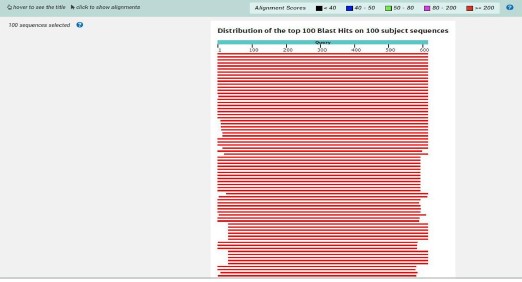
DNA sequencing: The graphical summary after alignment of the DNA sequence of the isolate GR-3E is shown in Fig. 3.
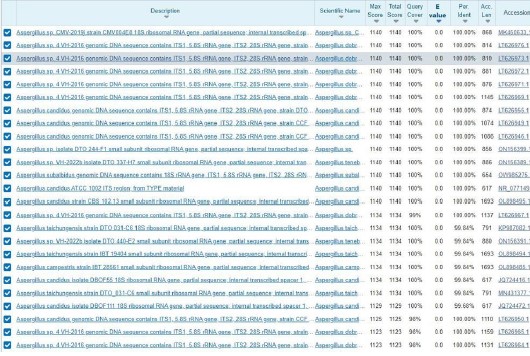
Sequence alignment using BLAST: Figure 3 indicated that the GR-3E isolate sequence aligned with 100 sequences deposited in the composite biological database of National Center Biotechnology Information (NCBI). Analyzing Fig. 4 showed that the GR-3E isolate sequence was 100% identical to Aspergillus candidus.
|

|
|
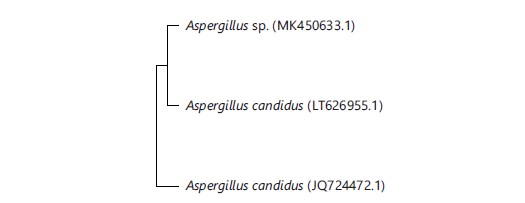
Phylogenetic tree: The phylogenetic tree constructed showed the relationship between the isolates from this study and other fungal isolates on Genebank. The phylogenetic analysis showed that Aspergillus candidus is closely related to the fungal isolates obtained from the goron tula as presented in Fig. 5.
Despite the numerous health benefits of goron tula, a lot of people eat mould infested goron tula without knowing the health implications of these moulds associated with goron tula and the traditional methods of identification are slower and less accurate than molecular methods, which offer excellent specificity in differentiating between fungal species and subspecies11. A distinct barcode for the characterization of various fungal isolates up to the species level is provided by molecular identification techniques based on complete fungal DNA extraction12. This study revealed the identity of the previously unknown fungal cultures to be Aspergillus candidus. This is the first documented instance of Aspergillus candidus functioning as a phytopathogen on goron tula fruits.
The fungus Aspergillus candidus obtained from this study belongs to the division Ascomycota, class Eurotiomycetes, order Eurotiales and family Aspergillaceae. A study conducted by Taligoola et al.13 indicated that this Aspergillus species is one of the most commonly seen moulds in cereal grains and flour. Aspergillus candidus produced a lot of dust on goron tula fruits and according to Krysinska-Traczyk and Dutkiewicz14 the concentration of A. candidus conidia in grain dust can reach hazardous levels and has been suggested to contribute to the development of the so-called organic dust toxic syndrome.
|
|
Aspergillus candidus is a fungus that lacks pigmentation meaning it is a white-spored species according to Flannigan et al.15, it is recognized as a pollutant of many different things, including foods, goods and the interior environment.
In this study, a new record of A. candidus causing fungi infection on goron tula fruits is provided.
It implies that Aspergillus candidus has been found to be associated with post-harvest rot in Azanza garckeana fruit. However, molecular characterization through sequencing of the Internal Transcribed Spacer (ITS) regions was effective in the identification of the specific Aspergillus spp., which is A. candidus fungus causing post-harvest losses in Azanza garckeana fruit. The research was limited to isolation and identification of fungi associated with Azanza garckeana using molecular and traditional methods. It is also recommended that more studies be done to determine the possible preservative(s) that can be efficiently used to combat fungal contaminants. Also, there is need for hygienic and proper handling of the fruits during harvest and processing.
CONCLUSION
The Azanza garckeana is a noteworthy native fruit tree with multiple uses in Nigeria. Azanza garckeana has a great deal of potential for application because of its mineral compositions and anti-nutritional qualities. The use of A. garckeana needs to be encouraged since it boosts fertility and will lessen the demand for artificial fertility-enhancing drugs. According to this study, Aspergillus candidus is one of the fungi that cause post-harvest rot in Azanza garckeana fruit. Thus, fruit should be thrown away if its color or flavor changes in a way that could pose a health risk to people. Based on identification utilizing phylogeny and morphology, we presented a new record of Azanza garckeana fruits disease in this study. Further studies are required to elucidate the primary inoculum and the inoculum source of the disease.
SIGNIFICANCE STATEMENT
Azanza garckeana, a female health remedy, has gained popularity due to its health benefits. Despite its numerous benefits, the consumption of infected seeds poses health risks. This study utilizes molecular methods to identify the fungi linked to A. garckeana seeds. Azanza garckeana is an essential export good and its fungal influence threatens farmers, consumer health and the economies of producing nations. The study of fungal ecology typically aims to determine the species composition and structure of fungal communities. This research will advance our understanding of the fungal diseases associated with A. garckeana and promote its use as a natural remedy for a number of health conditions, including lubrication, low libido, vaginal odor and infertility. The study found isolate GR-3E to be 100% identical to Aspergillus candidus.
ACKNOWLEDGMENTS
The authors wish to express gratitude to the sponsor of this publication, Dr. Ikechi Ahamefula Nwogu, University of Port Harcourt Choba Nigeria and the Department of Plant Science and Biotechnology for providing equipment and facilities for the experiment.
REFERENCES
- Yusuf, A.A., B. Lawal, S. Sani, R. Garba, B.A. Mohammed, D.B. Oshevire and D.A. Adesina, 2020. Pharmacological activities of Azanza garckeana (goron tula) grown in Nigeria. Clin. Phytosci., 6.
- Ochokwu, I.J., A. Dasuki and J.O. Oshoke, 2015. Azanza garckeana (Goron Tula) as an edible indigenous fruit in north eastern part of Nigeria. J. Biol. Agric. Healthcare, 5: 26-31.
- Bioltif, Y.E., N.B. Edward and T.D. Tyeng, 2020. A chemical overview of Azanza garckeana. Biol. Med. Nat. Prod. Chem., 9: 91-95.
- Mojeremane, W. and S.O. Tshwenyane, 2004. Azanza garckeana: A valuable edible indigenous fruit tree of Botswana. Pak. J. Nutr., 3: 264-267.
- Djarova, T., T. Kudanga and F. Chirimba, 1998. Quantitative determination of proteins, lipids and ascorbic acid in indigenous legumes and fruits of Zimbabwe. Trans. Zimbabwe Sci. Assoc., 72: 7-10.
- Jacob, C., Z. Shehu, W.L. Danbature and E. Karu, 2016. Proximate analysis of the fruit Azanza garckeana (“Goron Tula”). Bayero J. Pure Appl. Sci., 9: 221-224.
- Maroyi, A., 2017. Azanza garckeana fruit tree: Phytochemistry, pharmacology, nutritional and primary healthcare applications as herbal medicine: A review. Res. J. Med. Plants, 11: 115-123.
- Ikechi-Nwogu, C.G., B.A. Odogwu and I.K.G. Uzoka, 2022. Fungal presence in cosmetic facial powder. Asian J. Appl. Sci., 15: 47-52.
- Snowdon, A.L., 1990. A Colour Atlas of Post-Harvest Diseases and Disorders of Fruits and Vegetables: General Introduction and Fruits. CRC Press, United States, ISBN-13: 9780723416364, Pages: 302.
- Saghai-Maroof, M.A., K.M. Soliman, R.A. Jorgensen and R.W. Allard, 1984. Ribosomal DNA spacer-length polymorphisms in barly: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci., 81: 8014-8018.
- Liu, D., S. Coloe, R. Baird and J. Pedersen, 2000. Application of PCR to the identification of dermatophyte fungi. J. Med. Microbiol., 49: 493-497.
- Landeweert, R., P. Leeflang, T.W. Kuyper, E. Hoffland, A. Rosling, K. Wernars and E. Smit, 2003. Molecular identification of ectomycorrhizal mycelium in soil horizons. Appl. Environ. Microbiol., 69: 327-333.
- Taligoola, H.K., M.A. Ismail and S.K. Chebon, 2004. Mycobiota associated with rice grains marketed in Uganda. J. Biol. Sci., 4: 271-278.
- Krysińska-Traczyk, E. and J. Dutkiewicz, 2000. Aspergillus candidus: A respiratory hazard associated with grain dust. Ann. Agric. Environ. Med., 7: 101-109.
- Flannigan, B., R.A. Samson and J.D. Miller, 2011. Microorganisms in Home and Indoor Work Environments: Diversity, Health Impacts, Investigation and Control, Second Edition. 2nd Edn., CRC Press, Boca Raton, Florida, ISBN: 9780429147234, Pages: 539.
How to Cite this paper?
APA-7 Style
Ikechi-Nwogu,
C.G., Egere,
M.U. (2024). Macroscopic and Molecular Approaches for Identification of Fungi Associated with Azanza garckeana. Asian Journal of Biological Sciences, 17(3), 324-330. https://doi.org/10.3923/ajbs.2024.324.330
ACS Style
Ikechi-Nwogu,
C.G.; Egere,
M.U. Macroscopic and Molecular Approaches for Identification of Fungi Associated with Azanza garckeana. Asian J. Biol. Sci 2024, 17, 324-330. https://doi.org/10.3923/ajbs.2024.324.330
AMA Style
Ikechi-Nwogu
CG, Egere
MU. Macroscopic and Molecular Approaches for Identification of Fungi Associated with Azanza garckeana. Asian Journal of Biological Sciences. 2024; 17(3): 324-330. https://doi.org/10.3923/ajbs.2024.324.330
Chicago/Turabian Style
Ikechi-Nwogu, Chinyerum, Gloria, and Mirabel Ufuoma Egere.
2024. "Macroscopic and Molecular Approaches for Identification of Fungi Associated with Azanza garckeana" Asian Journal of Biological Sciences 17, no. 3: 324-330. https://doi.org/10.3923/ajbs.2024.324.330

This work is licensed under a Creative Commons Attribution 4.0 International License.



