Unveiling the Dynamics: A Comprehensive Review of Biological Membrane Kinetics During Cell Division
| Received 19 Jul, 2024 |
Accepted 21 Sep, 2024 |
Published 31 Dec, 2024 |
A cell division is a series of events that occur tandemly during cell growth. Most of the cell cycle is involved in the phase known as interphase, during which it matures, copies its chromosomes and prepares to divide. Following interphase, the cell completes mitosis and divides to complete offspring lineage. As a result, each of the daughter cells that form enters its interphase to begin a new cycle of the cell. During the cell division, eukaryotic cells undergo complicated and well-coordinated changes to their membranes and cytoskeleton. Organelles such as the nuclear envelope, endoplasmic reticulum and Golgi apparatus must be destroyed or altered, distributed and then rebuilt to complete the cell cycle. The precise distribution of cytoplasmic material and an appropriate number of organelles was also required between daughter cells. As cell division progresses, the plasma membrane that covers this mixture of cytoplasm and organelles is prone to alter. Resident lipids in these distinct membrane compartments exert their considerable influence on the division process. The cell splits into two daughter cells along with the two copies of the genetic material. During cell division, cell-cycle proteins have a role in both the regulation and maintenance of the cell cycle in eukaryotic cells. They regulate transitions between interphase, mitosis and cytokinesis, the three phases of the cell cycle that result in cell division and replication. Therefore, cells have developed sophisticated organelle inheritance mechanisms to permit efficient intracellular organelle sharing. To completely understand how the remodeling of various organelles is interconnected and how changes in organelle shape and function affect the process of cell division, it will be obvious to combine this information in the current context. The study aimed to review and synthesize current knowledge on the kinetics of biological membranes during cell division.
| Copyright © 2024 Bhattacharya et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
All living organisms are made up of cells, which are the smallest units capable of independent life. Cell division is a dynamic process in which the parent cell is divided into daughter cells and the cellular components of the parent cell duplicate properly1. All eukaryotes share common unicellular predecessors and many parts of the cell cycle mechanisms are conserved2. Multicellularity requires the emergence of mechanisms that coordinate cell growth and proliferation across growing organs with the adoption of specialized cell fates3. There are two different types of cell division, the first one being vegetative division known as mitosis, in which each daughter cell replicates the parent cell. Meiosis is the second one to generate four haploid daughter cells which are also known as reductional division. Prophase is the first step of mitosis in which rearrangement of microtubules occurs and the DNA molecules are condensed gradually to form chromosomes4. The disassembly of the nuclear envelope is the feature of prometaphase and the alignment of chromosomes in the metaphase plate is observed during the metaphase stage in which the kinetochores are attached to the chromosome arms and are connected to opposite poles of the spindle fiber through the microtubules5. After completion of the metaphase stage when the spindle assembly checkpoint arrived, segregation of chromatids started in the anaphase stage6. Chromosomes are separated from each other and move towards the opposite pole. In telophase, chromosome segregation is mediated by the mitotic spindle7. In cytokinesis, the midbody is a structure formed in mammalian cells by the condensation of mitotic spindles8. According to Mierzwa and Gerlich9, this process is called cytokinetic abscission where two daughter cells are separated by cleaving of the midbody. Cell division is completed by cytokinetic abscission, which results in the physical cutting of the intercellular bridge (ICB) that connects the daughter cells10. At the time of division, to accurately segregate the genetic material from the parent to the daughter cells, a high level of both spatial and temporal control is needed inside the cell.
After completion of the cell division, the newly formed daughter cells have to remain functionally active as they need to process the nutrients for pursuing their metabolism so they can produce proteins, DNA and other components as per their requirements for their survival and growth. Most of the cellular organelles are newly modified during the cell division procedure. In daughter cells, de novo synthesis of the cell organelles is not possible due to the requirement of time and resources and it also causes inequality of organelles in the daughter cells11. The division of cell organelles is a complex issue for a dividing cell as there are so many criteria like size, numbers, shape and locations. The organelles are also following different methods for division. Some of them are segregated into pieces to the cytoplasm and then reaggregate after the division of the nuclear membrane and Golgi bodies12. Some organelles like mitochondria may change their external structure and retain some of their stochastic inheritance but they never segregate and reaggregate12. Not even all the time the partitioning is correct, in some cases, such issues need to be solved by different methods and those are also different from organelle to organelle13. Therefore, it is clear that organelles require their mechanisms to maintain their characteristics. A number of proteins are involved in both cell division and organelle biology. Membrane-bound organelles contain the proteins that are recognized by the midbody for proteomic analyses and mitotic failure screened by RNA interference14-19. Scientists made a whole catalog for different types of proteins of the membrane compartment that have marked effects in the cell division process and also in organelle biology. Several proteins in this catalog are known to scientists but they are not deeply studied in this cell division context and some of the proteins are very less known20-23. In this review, several examples are taken from different organisms mainly highlighting the mammalian cell division steps to lipid dynamics. The specific objectives of this review are to review and synthesize current knowledge on the kinetics of biological membranes during cell division, focusing on the role of lipid composition and membrane compartmentalization; explore the mechanisms underlying the remodeling, distribution and inheritance of organelles during cell division, emphasizing their interconnection with membrane dynamics and analyze the regulatory roles of cell-cycle proteins in coordinating membrane and cytoskeletal changes throughout the phases of cell division, including interphase, mitosis and cytokinesis. Here, the key mechanisms and processes that regulate membrane kinetics during cell division, such as membrane trafficking, lipid and protein rearrangements and the involvement of cytoskeletal elements are highlighted. In essence, the review would aim to provide a comprehensive summary of how biological membranes are involved in and affected by cell division, integrating existing research and identifying directions for future work.
How do lipids affect the division of cells? Lipids are essential for cell function in processes like organ morphogenesis, cell differentiation and organ proliferation, all of which are closely related to cell cycle development24. Lipids are involved in numerous biological activities and makeup a major portion of membranes. Dramatic changes in cell structure trigger cell division. The cytoskeleton and lipid remodeling, as well as local protein complex recruitment and membrane trafficking, are required for each phase of cell division25. Except for sterols, lipids are significantly smaller than proteins and typically consist of a polar head group facing the membrane exterior and one or more hydrophobic carbon side chains facing the membrane lipid bilayer interior26. As per head membrane protein, nearly 50-100 of them are present in the cellular context27. The length and saturation of hydrocarbon side chains might vary, resulting in complex and diverse lipidomes when combined with different head groups28. Lipids play a variety of activities in the cell including structural and signaling functions. The considerable energy requirements for the synthesis and maintenance of the wide variety of lipid molecules have been retained throughout species, demonstrating the crucial functions of lipids in cellular physiology27. At the molecular level, it is not clear why the cells devote their most vitality to manufacturing and keeping the lipid substances and how they restrain them. However, it is well known that cells communicate through complicated and different lipidomes28. Different percentages of lipids are present in the different organelle29, which causes a difference in the interactions between protein and lipid which affects the fluidity thickness and other characteristics of the membranes30. As mentioned by Scorrano et al.31 the contact sites of the membranes are known as the main functional sites of the cells in which the lipids can alter between organelles. According to Muro et al.32, lipids are difficult to analyze since they are not generated iteratively like proteins or nucleic acids and hence cannot be systematically deleted or tagged.
Different lipids confined to the structures related to the division like the midbody and also the cleavage furrow can imply cell division33-35. In biological membranes, phospholipids are distributed asymmetrically between the inner and outer leaflets of the lipid bilayer36. Membrane lipids are hydrolyzed by phospholipases to produce a variety of cellular mediators. Several phospholipase-derived substances, including diacylglycerol, phosphatidic acid, inositol phosphates, lysophopholipids and free fatty acids are essential for signal transmission throughout plant growth, maturity and stress responses37. The faithful duplication of cellular components depends on the synchronization of metabolism and cell cycle development. A misaligned regulation would cause cells to have an excessive or insufficient amount of membrane surface, which would cause anomalies in cell size. Prior to cell division throughout the regular cell cycle, cells must manufacture more membrane phospholipids38. Membranes are interrelated with lots of cell division proteins27. Few membrane proteins are integral and they can expand the lipid bilayer. Whereas others are linked peripherally with the membranes by the domains attached with lipids like phosphoinositide are linked or attached with pleckstrin homology (PH) domains. Lastly, a few proteins of cell division are attached to the lipid moiety after translation, so these are mentioned as lipidated. Cells that are in dividing mode can control their lipid constitution and placement in them properly39. Echard40 suggested that the role of phosphoinositide in cell division is well understood in directing the dynamics of actin and the development of the midbody. There is a complicated interconnection between the lipids and proteins of the cell during cell division. In this process, how both of these molecules are connected and interact together is the main thing to decipher.
|
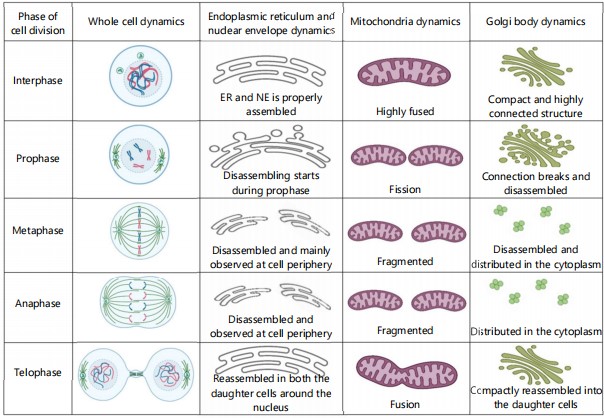
Organelles as the building constituent of the cell: The cells give any organism shape and structure as well as perform a variety of other duties. For conducting different functions, a cell has several structures inside it named cell organelles which have individual functions to perform as well as either work singly or some of them work together41. If any one of them is not performing its function properly then definitely the whole body will be disturbed41. Each organelle has its own structural and functional changes during the cell division process. There are so many cell organelles bounded by the selectively permeable plasmalemma. In the cell cytoplasm, the cell organelles are present to perform their specific function. Some of them are membrane-less, some are single-membraned and some are double-membraned42. The nucleus is the largest double-membraned organelle of the cell, which mainly has all the genetic information of the cell. A network of fluid-filled membranous channels constitutes the endoplasmic reticulum of the cell43. They are the cell transportation system that is responsible for the movement of materials around the cell. Next is the mitochondria, which is the membraned structure known as the powerhouse of the cell44. Plastids are large membrane-bound organelles that contain pigments that are present in the plant kingdom45. Ribosomes are membrane-less cytoplasmic organelles found close to the endoplasmic reticulum46. Its primary function is to synthesize protein. Another important cell organelle is the Golgi apparatus, which is membrane-bound and helps in protein sorting47. Centrosomes are organelles comprised of centrioles and found only in animal cells48. Another tiny organelle is the lysosome which helps in cellular digestion49. The peroxisome is the membrane-bound organelle having several reducing enzymes50. These are the major cell organelles that have different roles in cell functioning and undergo several changes during cell division (Fig. 1).
Control of the cell cycle involves organelles: A crucial component of the regulatory network that controls the progression of each phase in the proper order is the cyclin and Cyclin-Dependent Kinases (CDKs). Additionally, both the CDK-cyclin complex’s targets and the transcriptional events that follow are evolutionarily conserved51. Stimulation of the main cell division kinase CDK1 is the first step of eukaryotes cell division. Then, auxiliary kinases such as Aurora A, Aurora B and Polo-Like Kinase 1 (PLK1) help in the division phase entry as well as perform a vital role in the division. It is generally believed that the functional and structural aspects of the cell cycle are primarily associated and connected with the cell nucleus52. Plant cyclins are unique in that they are diverse, across over 50 homologs in arabidopsis53. This diversity is thought to be a reflection of the various environmental factors that influence the course of the plant cell cycle as well as the specific cyclin functions carried out by various cell types3. The cell cycle’s one-directional character is retained by targeted proteolysis of the major proteins. Phosphatases return cells from the dividing stage to the interphase stage once they have completed cell division54,55. Recent research suggests that synchronized alterations in the structure and positioning of intracellular organelles occur during mitosis.
Spatiotemporal localization of kinases and phosphatases during division: At the time of division, the CDK1 scattered throughout and Aurora A, Aurora B and Polo-Like Kinase 1 (PLK1) localize vigorously which helps in the error-free spatiotemporal control of cellular renovation. One of the master kinases in mitosis, PLK1, is involved in a variety of processes, including chromosomal segregation, spindle assembly checkpoint, cytokinesis and mitotic entry. Understanding how PLK1 localizes in a particular area of cells is crucial to analyzing the function of PLK1 in mitosis56. Some supplementary regulator kinases help to control particular incidents in kinetochores, like-MAST-L, MPS1 and serine/threonine-protein kinases haspin. The PLK1 is a kinetochore-localized protein that is crucial for the spindle assembly checkpoint and chromosomal segregation. However, still it is unclear how PLK1 localizes at the kinetochore56. Kinases are very important as they build the mitotic state by changing different protein-protein relationships and actions. Combes et al.57 stated that the accuracy of cell division can be increased by the site-specific events of phosphorylation and dephosphorylation. In the spindle midzone, the placement of Aurora B makes an inclination toward its activity and influences the substrates to detach when the spindle extends58. There is a ‘ruler-based’ model of mitotic exit which explains that on lagging chromosomes Aurora B-dependent remembrance of condensins permits continuing chromosome loosening and nuclear rebuilding59. According to Wang et al.60, the same inclination to Aurora B is there around the centromeres, from which it may conclude that diffusion gradients of a place may be a usual donor to the spatiotemporal controller of mitotic events. Mitosis-specific phosphorylations are separated by PP1 and PP2A, which are proteins that phosphatases in the time way out from the division of the cell54,55,61. The CDK1 phosphorylation inhibits PP1 activity, as does MAST-L-dependent phosphorylation, which is followed by the triggering of endosulfin, as indicated by ENSA, a PP2A inactivating protein51,62-64. The number of regulatory subunits is much more than those works like adaptors for the enzymes that obstruct our knowledge about biological matters65. The Repo-man and Ki67 are examples of PP1 adaptors that set up PP1 holoenzymes which create circumstances that help the cell to exit the cell division process like the reappearance of the nuclear envelope and reformation of chromatin66-69. The B55 and B56 are PP2A governmental B-type components with specific features for the mitotic exit, such as assisting in the reconstruction of nuclei, nuclear envelope and Golgi bodies61,64,70.
Controlled proteolysis during cell division: In the case of the cell division process, the regulated proteolysis mechanism plays a significant role. The CDK regulates the progression of the cell cycle at distinct stages but regulates proteolysis in the case of cyclins71, sister chromatid cohesion proteins72 and major mitotic kinases73-74 to secure the unidirectional progression. Recent research indicates that there may be numerous different ways in which the cell cycle-independent metabolic oscillations connect with the machinery involved in the cell cycle and certain biosynthetic pathways have temporally varying activity patterns throughout the cell cycle75. The cell cycle regulators are knocked down in proper time by ubiquitin ligases which keep forward progress. So, without these ligases, the progression in the right direction is not possible. For example, at the time of division, the anaphase conducting complex/cyclosome (APC/C) is present and the left of the process follows. The SKP, cullin and the F-box-carrying complex are all present to keep the process in check76. Separation of chromatid and exit from the division state is carried out by regulated proteolysis. Direct proof for the value of proteolysis in the inheritance of organelles is very limited but it is proved that proteolysis indirectly controls the organelle dynamics due to the difference in the kinases and phosphatases levels. According to Sivakumar and Gorbsky77, the role of APC/C is important as it suggests a spatiotemporal activity that ensures the proper settlement of cellular reorganization during mitosis. Horn et al.78 mentioned that mitotic levels of DRP1 are controlled by APC/C. This is the main ubiquitin ligase that is needed for the fission of mitochondria and organellar proteostasis those combined with cell cycle progress.
Dynamic behaviour of nuclear envelope and endoplasmic reticulum during cell division: The endoplasmic reticulum (ER) creates a complex connected network of membrane layers and tubules. The endoplasmic reticulum is a polygonal network of linked tubules and layers that spans the whole eukaryotic cell and is next to the nuclear membrane79. The nucleus is surrounded and enclosed by ER membranes so there is a formation of a bi-membraned nuclear envelope (NE). The main role of this nucleus membrane is to give protection to the chromatin. Throughout the luminal and membrane sections of the ER, numerous nascent proteins are folded and matured. Despite the fact that the ER contains a number of elements that encourage protein folding, many proteins end up being destroyed because they are unable to fold and assemble correctly80. As reported by Güttinger et al.81 and Champion et al.23 after the NE, a series of intermediate filaments known as lamin like lamin A/C and lamin B create the nuclear lamina. This nuclear lamina gives mechanical rigidity to the nucleus and also helps the NE to organize the chromatins properly82. In this bi-membraned NE, massive macromolecular structures called nuclear pore complexes (NPCs) aid in transit throughout the nuclear membrane83. It joins the nucleoskeleton and cytoskeleton complexes to operate and connects the cell's nucleus and cytoskeleton84. The ER is a vital organelle that cannot be generated from scratch thus, it must be split into daughter cells before cell division79. The exterior nuclear membrane is always in contact with the ER, but the interior nuclear membrane (INM) is biochemically different from the outer one because it has its own set of proteins that interact with the lamina, histones and the proteins that are held together with chromatin85,86. The NE disintegrates in higher eukaryotes during the open cell division process and the destiny of the NE along with its components has piqued researchers’ curiosity, especially in metazoans87. The INM is a highly specific membrane segment with several native intermembrane proteins that take an essential part in nuclear engineering, such as coupling the INM to chromatin and the nuclear lamina’s intermediary filament network, transcription control and signal transduction80,88. It can be observed distinct morphologies in ER and NE during the interphase. The NE is a disjointed and flexible platform that affects gene expression and chromatin organization. In plant mitosis, NE offers a structural and regulatory framework. However, it must be rebuilt after each cell division89. The cisternal and tubular-reticular architecture of the external ER is created by membranous sheets and numerous morphometric proteins that make up the NE. The Reticulon (RTN1-RTN4) and Receptor expression-increasing proteins (REEP1-REEP6) are proteins that stabilize majorly curved ER tubules and the curved edges of sheets90,91. As mentioned by Nixon-Abell et al.92 and Schroeder et al.93 super-resolution microscopy reveals more morphological differences, such as ER matrices and reactive nanoholes. Microtubule cytoskeleton forms the interphase ER and during division, many modifications take place to allow the remodeling of ER94,95.
Nuclear envelope breakdown is brought at the commencement of mitosis: Nuclear envelope breakdown (NEBD), which entails the loss and repositioning of the NE's structures and components, is essential for open mitosis. In the late prophase, NEBD in plants occurs relatively early than in animals96. The CDK1 relying on phosphorylation of INM-chromatin and INM-lamina binds the nuclear membrane to chromatin and aids in the redistribution of nuclear membrane proteins to the mitotic ER at the time of a cell’s mitotic entrance23. The CDK1 and PKC phosphorylate the lamins which disintegrate the nuclear lamina and help to free the NE from the nucleus97,98. As reported by Bahmanyar et al.99 phospholipid metabolism is spatially controlled when phosphatidic acid is inactivated and phosphatase lipin restricts de novo ER membrane formation to aid in the breakdown of the nuclear membrane. After the NE is disassembled, each NE and ER characteristics are destroyed. The envelope protein molecules are split into daughter cells using ER elements. Audhya et al.100 reported in the case of Caenorhabditis elegans that nuclear membrane disassociation is carried out by the reticulons and also suggested that a more tubular morphology is formed from the sheet-like NE which aided in the demembranation. Dynein creates pulling forces that aid in demembranation by the generation of NE invaginations in prophase101-104. As observed by Mori et al.105 rapid polymerization of an F-actin layer at the nuclear border aids physical breakdown of the NE in starfish oocytes. Cyclin-Dependent Kinases (CDKs) have primary control over cycle progression, making them important conserved regulators. Animal CDK1, Saccharomyces cerevisiae Cdc28 and Schizosaccharomyces pombe Cdc2 are all closely related to plant CDKA, which is active throughout the cycle. Plants only have a second form of CDK, named CDKB1 and CDKB2, which is active from the S to M phase96. But in mammalian cells, the contribution of actin in mediating NE disassociation is not clear and this will be a major area of study in the future106. At the start of mitosis, the rapid disassembly of NPCs is carried out by the phosphorylation of nucleoporins (NUPs)107. The transmembrane NUPs enter the mitotic ER and the other NUPs are liberated into the cytoplasm108,109. Current studies show that in early mitosis, the nuclear pore knockdown and the subsequent disassociation of the nuclear membrane are the results of the transient localization of PLK1 to NPCs. The PLK1 and CDK1 act together and hyper-phosphorylate NUP98 and NUP53. These take the main step toward the separation of the key interconnection of NUPs and the breakdown of the nuclear pore56,107,110-112. At the start of mitosis, this kind of phosphorylation-induced separation of inter-NUP interactions is the major cause of the disintegration of nuclear categorization and disassociation of the NE.
Division results in morphometric changes to the ER: The morphology of the ER in the mitotic phase is not clearly known but in interphase ER shows various tubulo-reticular and cisternal subdomains. The outer nuclear membrane (ONM) as well as the inner nuclear membrane (INM), which creates the nuclear envelope (NE) is continuous with the ER80. Live cell imaging of mitotic cells reveals that extended curvilinear membranes are present in the ER membrane with a cisternal arrangement113,114. Volume electron microscopy approaches reveal the tubular-reticular and peppered sheet structure formation of ER115-116. During the division, the ER-forming proteins REEP3 and REEP4 form the tubular architecture. The cell type determines how much each morphology predominates116. The nucleoplasm and cytoplasmic proteins that interact with the envelope are essential to its completion, as are timing and location96. Schweizer and Maiato117 reported that in dividing cells the peripheral ER is not attached to the spindle and it forms a barrier to make possible the exit of other organelles. The ER morphology can be affected by mitotic phosphorylation of ER morphometric proteins by stopping LNP multimerization118-119. Because of these events, the connection between the ER and microtubules is affected to stop the ER from assembling with spindle assembly120. As observed by Schlaitz et al.121 REEP3 and REEP4 are also critical for concentrating the ER at spindle poles, preventing them from forming early associations with chromatin and resulting in cell division error. The attachment of ER with spindle poles and astral microtubules controls the ER inheritance122. Therefore, it is important to understand definitely through which process the spindle controls the inheritance of ER without letting these organelles hinder chromosome segregation123.
Regeneration of nuclear enclosures: The NE reappears in late anaphase and telophase when the daughter nuclei are connected and surrounded by ER membranes23,124,125. Unlike animal cells, the multiplying plant cells build a new cell wall to finish their division. It is necessary to synchronize the division plane because the wall inhibits later correction. The wall inhibits the swelling that happens during animal cell division. As a result, the membrane plays a significant role in the formation of the two plasma membranes as well as the movement of vesicles that carry the building blocks for the new wall96. The switch from tubular to sheet-like architecture is a frequency-limiting phase in NE rebuilding as evidenced by the fact that overexpression of ER tubule-shaping proteins hindered the prior process126. The chromatin-associated protein molecule barrier-to-autointegration factor (BAF) forms a crosslinked chromatin interface where the ER membranes are assembled127. Reversible disintegration of the interphase lamina may govern NE deconstruction and rebuilding during mitosis, based on the physical features of the interphase lamins and their activities throughout the mitosis. The NE reappearance requires reversal of the mitotic state and PP1 and PP2A play crucial roles in linking nuclear reorganization and chromatin decondensation. Thus, the absence of these PP1 and PP2A affects the reorientation of NE. The PP2A governing component B55 is necessary for NE reconstruction during mitotic departure61. In timely nuclear membrane reorientation, lamin rearrangement, NPC reorganization and the re-employment of INM-chromatin need PP1 by its targeting subunits PNUTS128, AKAP149129, Repo-Man67 and Ki67130,131. The NE reappearance in late-segregating chromosomes can be delayed by Aurora B which suggests that re-memorization is controlled spatially by the midzone Aurora B gradient132. This type of Aurora B-dependent spatial regulation of nuclear membrane reassembly may happen even in human cells. The NE governs macromolecule interchange between the nucleus and the cytoplasm and may have a role in the interphase chromosomal architectural organization133. However, modern research suggests that rather than membrane generation, it regulates the presence of NPCs and the nature of the NE134. The NPCs that have been disassembled at the end of mitosis are reassembled by incorporation into NE which has previously been secured by the formation of the membrane around a pre-existing pore structure. The nuclear envelope (NE), which serves as both a physical barrier partitioning the nucleus from the cytoplasm and a gatekeeper for selective transport between the two, is essential to the structure of eukaryotic cells. In contrast, the NE breaks down during open mitosis to enable spindle formation and chromosomal segregation, which causes the nuclear and cytoplasmic soluble portions to mix135. The nuclear lamina, a fibrillar protein meshwork (typically 10-20 nm thick) that lines the nucleoplasmic face of the inner nuclear membrane appears to be connected to pore complexes. Relating live-cell electron tomography illustrates that as anaphase continues, small gaps in the aperture of NE sheets expand to gain the morphological features of NPCs136. Transient intracellular calcium elevations may influence the advancement of mitotic cells into anaphase. It is concluded that the NE is assembled around one pre-existing aperture at this moment. The NPCs can also be incorporated into the centre of the nuclear membrane when the nucleus increases, simulating NPC integration in the interphase. According to Otsuka et al.137 through the INM and extrusion of a pre-existing body NPCs can form at the time of mitotic exit as revealed by tomographic approaches. In fact, at the time of mitotic exit, the lamina and the INM proteins are formed by nucleoporins138. The leftover holes in the NE are sealed by membrane fusion which is the last event in nuclear regeneration. Point fusion in ER membranes allows NE to expand to accommodate decondensed chromatin and plug holes in the two-layered NE, splitting the internal and external nuclear membranes associated at these places where a specialized fusion event is required. The ESCRT-III mediates this process (annular fusion), which is responsible for ontologically identical membrane filtration events in several other cellular activities, for the assignment of the endosomal sorting complex10,139,140. In light of the interphase lamins' physicochemical features and simultaneous disintegration of their behaviour during mitosis, the NE deconstruction could be regulated by the lamina and the process of mitotic rebuilding. The CHMP7 is an ESCRT-III component found in the ER that transiently crosses target places during annular fusion by interacting with the INM protein LEM2141,142. It is well known that the ESCRT-III complex causes the intercellular bridge to tighten and scissor in several animal cells10. Elements of the ESCRT-III are allocated to close the leftover holes in the NE, allowing nuclear fragmentation139,140. Gaps in the NE are formed when membranes enclose spindle microtubules and the microtubule-severing enzyme spastin is allocated by ESCRT-III to associate microtubule disassembly with membrane sealing140. However, the connection between the mitotic reassembly of NPCs and this process is still unknown143-145. The discovery that this pathway exists in archaea shows that this mechanism has evolved to play a function in cellular division146.
Mitochondria during cell division: Mitochondria are bi-membranous organelles that serve as the cell’s primary energy producers. Plant mitochondria are pleomorphic, active organelles. The higher plant chondroma (all mitochondria in a cell) is usually made up of a large number of morphologically distinct mitochondria147. It is suggested by Labbé et al.148 that continuous fusion and fission happen inside the mitochondria for adaptation to the energy requirements of the cell. Mitochondria contain their independent genomes that are duplicated in a separate growth and division cycle from the cell cycle, something that the other organelles lack. The genes that regulate mitochondrial fusion in plants are unknown; most of the genes reported to be associated with fusion in other organisms have no plant homologs147. Therefore, a dividing cell must finish two tasks at a time including mitochondria division, which firstly optimizes energy production at the time of division and secondly relocate mitochondria between the daughter cells. Mitochondria are extremely dynamic organelles that experience fission, fusion and transport events during various cell cycle stages. Notably, many kinases phosphorylate the essential mitochondrial components and promote mitochondrial fragmentation during mitosis to enable efficient mitochondrial dispersion and transmission to two daughter cells149. As proposed by Mishra and Chan13 mitochondria are united throughout the G1 phase, stretched during the S phase, divided during G2-M transition and shattered during division. Dynamin-related GTPase proteins (DRPs) control mitochondrial fission-fusion cycles. Labbé et al.148 proposed that mainly DRP1 controls the fission and the distribution of mitochondrial DNA at ER-mitochondria contact sites. Mitofusin 1 and Mitofusin 2 (MFN1 and MFN2) are DRPs that control mitochondrial outer membrane merging, while DRP OPA1 controls mitochondrial inner membrane merging. According to Fenton et al.150, the morphology of the mitochondria can change depending on a number of cellular signals. Increased intracellular calcium and mitochondrial depolarization are two stimuli that are frequently employed to modify mitochondrial network architecture. The Aurora A through the small GTPase RalA and CDK1 activates and phosphorylates DRP1 during G2-M, which results in higher mitochondrial fission151,152. The mitochondrial disintegration mechanism, on the other hand, is highly preserved147. This may be coming together by the ubiquitylation and breakdown of MFN1 and MFN2153. Higher plants’ mitochondria are reported to differ significantly from other eukaryotes in a number of ways154. The DRP1 is sumoylated and ubiquitylated in mitotic exit, causing its disintegration and restoration of mitochondrial fusion following cytokinesis78,152. According to Adachi et al.155, the activity of DRP1 is restricted by attaching to phospholipids and this incident suggested that other ways may also take part in controlling mitochondrial position during cell division. Dynamic mitochondrial partitioning may potentially contribute to cell fate determination. Mitochondria are involved in the production of various substances, including phospholipids, nucleotides and many amino acids, in addition to their critical roles in respiration and photorespiration147,149. As a result of the uneven splitting of stem cells, the more stem-like daughter cell maintains newer and more efficient mitochondria156. During the cell dividing process, Mitochondrial disintegration may aid in the stochastic transmission of mitochondria as suggested by Jajoo et al.157. Mitochondria are pushed and placed to the ends of dividing cells in Schizosaccharomyces pombe and re-disseminated soon before splitting in according to the partitioned cytoplasmic content. Mitotic kinesin KIF5B, coupled with MIRO1, attaches astral microtubules to transport mitochondria to the trench during the mitotic departure158. The MIRO1 and MIRO2 appear to operate as a connector between the mitochondria, microtubules and the actin cytoskeleton. The other of these connections can be made via the non-processive myosin drive unconventional myosin-XIX, which is transcribed by MYO19 and found in mitochondria’s outer layer159. During mitotic entry and exit, the control of mitochondrial-cytoskeletal interactions permits future studies to know how cytoskeletal targeting aids accurate inheritance patterns. The activity of particular cyclin-Cdk complexes determines cell-cycle progression. These complexes control several structural changes essential for cell division, including changes in mitochondria shape that allow for mitochondrial segregation across daughter cells21,160-162. Major avenues of cellular processes, including crucial roles in the regulation of immunity and mitochondrial quality control, appear to be influenced by mitochondrial division163. How cells regulate the availability of energy throughout the cell division mechanism is a vast and fascinating subject. Mitochondria are the main energy makers of the cell and they could aid cells to satisfy the high energy needs during the whole division process164. In addition to offering power for life, mitochondria can serve as a death trigger147. At the activation by Aurora A, the small GTPase RalA comes to the mitochondria. The degradation of RalA and RALBP1 demonstrates a regulatory link between the division mechanism and the cell’s power supply151.
Golgi apparatus disassembly and suppression of transport activity: To continue the equal inheritance at the time of cell division, the Golgi body must be disorganized and again reorganized in daughter cells as this is a single-copy organelle. Several extended piled cisternae formed an interconnected ribbon-like pattern in Golgi165. However, certain protozoa only have a solitary Golgi mini-stack within the cell and cytokinesis is purely in charge of controlling the mini-stack's division166. Outer Golgi reassembly and piling proteins (GRASPs) such as GRASP55 and GRASP65, play an important role in Golgi structure167,168. The Golgi apparatus in mammalian cells is made up of a highly organized parallel network of cisternae that form a layered architecture in the cell’s perinuclear zone166. Golgins are a class of coiled-coil proteins that interact with anchoring factors such as p115 to grab vesicles169,170. Golgins can be linked with GRASPs for making the structure of the Golgi. In proper controlling of biosynthetic and endocytic trafficking pathways, the Golgi has the main role at the time of cell division to regulate the remodeling program that certifies its disassembly and restoration. Golgi dispersion begins in the late G2 phase of the cell cycle. As the C-terminal-attaching protein, 1/brefeldin A ADP ribosylated substrate (CtBP1/BARS) breaks links between cisternae layers, the Golgi ribbon splits into solitary ministacks171,172. At the time of mitotic entry, GRASPs can be phosphorylated by CDK1, PLK1 and MAP kinases which conceal the trans-oligomerization and help in the Golgi ribbon unattached173-178. The Golgi apparatus is hypothesized to have a role in the production of complex carbohydrate frameworks that are coupled to a variety of cellular and secretory proteins and lipids, as well as the filtering of these proteins and lipids to their proper subcellular locations. The GRASP65 also stabilizes microtubules, which are required for interphase Golgi aggregation. Ribbon unlinking is caused by its phosphorylation and deactivation after mitotic entry, which subdues its attachment to the cytoskeleton179. The shifting of Aurora A to centrosomes is halted by blocking the unlinking of the Golgi ribbon, which also stops the mitotic entrance180. At the beginning of mitosis, the GC is rearranged to form an entirely new ribbon in each of its daughter cells. During mitosis, the GC is fragmented into vesicular/tubular clumps that are scattered in the cytoplasm until the infamous “Golgi haze” is created178. Elements that contribute to the fusing of Golgi membranes to generate cisternae and the following stacking of these cisternae to form stacks have been found in a number of investigations in recent years168. Cells keep monitoring the stability of this organelle to allow mitosis to continue if the correct lineage is feasible174,181. After the unlinking of the Golgi stack, the coatomer COPI helps the Golgi cisternae to vesiculate through persistent budding. Phosphorylation of golgins such as GM130 paused the vesicular fusion to inhibit the relation with p115182-184. Membrane fusion at the Golgi is also controlled by the Ubiquitin dynamics. For instance, Tang et al.185 and Huang et al.186 reported that HACE1, an E3 ubiquitin ligase, ubiquitylates the Golgi SNARE syntaxin 5 to halt its association with its corresponding SNARE BET1 and disrupts integration activities at the Golgi when mitosis begins. The VCIP135-dependent deubiquitination of this SNARE restores union, whereas phosphorylation of VCIP135 blocks this action in mitotic cell division187,188. The GRASP55, a GRASP65-related mammalian protein, is involved in the development of Golgi bundles168. The ATPases NSF and valosin-containing protein allow the disassociation of trans-SNARE complexes to maintain continuous cycles of vesicular integration processes. Mitotic phosphorylation of the VCP-adaptor p47 removes this protein from walls to stop VCP-dependent membrane addition in mitosis when mitotic phosphorylation of GM130 obstructs NSF-mediated union184,189. Small tubulovesicular clusters are developed through the inhibition of membrane fusion to form cytoplasmic Golgi ‘haze’ and this haze may be divisible into daughter cells by stochastic mechanisms178. The inheritance of these membranes is supported by a mitotic spindle and GM130-microtubule interlinkage190. Another theory proposes that the reorganization of Golgi parts into the ER is caused by a halt in transport191,192. During the G2–M transition, Golgi separation occurs in tandem with global cellular reintegration activities such as spindle formation. Following the reassembly of the Golgi complex, there is an issue with the mitotic breakdown in a cell-free system. In the first phase, cisternae are reconstructed from the ground up MGFs193. During mitotic entry, CDK1 phosphorylates GM130184 which detaches its connection with p115 and a nuclear localization signal is produced within GM130. Importin-alters its location and is deposited in Golgi membranes as a result of this nuclear localization signal, releasing it to inhibit TPX2-dependent spindle formation190. The Golgi restoration takes place mostly in between the anaphase and cytokinesis stages. The reformation of the Golgi cisterna, restacking of the cisterna, congression and fusion of piles to produce ribbons are all required for Golgi rejuvenation. The Golgi apparatus’s organized architecture is hypothesized to represent the need for these enzymes and the protein filtering mechanism to be partitioned to carry out a particular set of post-translational alterations and sorting activities. Metal ions have recently been shown to be essential for the Golgi complex to continue functioning properly194. The PP2A-dependent dephosphorylation of GM130 reconstitutes p115-dependent vesicular tethering and fusion195. With this, cisternal re-stacking is permitted by PP2A-mediated dephosphorylation of GRASP65196,197. Both radial and tangential microtubule-dependent mobility help to rebuild a Golgi string198. ‘Twin’ Golgi are formed in each daughter cell in telophase199. A shorter twin faces the cleavage trench, while the bigger twin faces the centrosome. In cytokinesis, the shorter twin moves around the nuclei to join its twin and recreate a single Golgi. The Golgi apparatus disperses into the farthest portions of the cell-cytoplasm in highly differentiated cells like the oocyte or nerve ganglion cell and so lacks its link to the centrosome in most situations. Goss and Toomre200 revealed that the smaller twin may deliver Golgi-derived particles to the midbody for abscission, although the rationale for this placement is unknown201. The ARF1 guanine nucleotide transfer factor GBF1 was recently found to be required for the movement and attachment of these two Golgi twins.
Actin and phosphoinositide clearance from the midbody: The reshaping of actin, change from a contractile band to a midbody band and the formation of the abscission region are all required for cortical entering. Many organelles in a wide range of plant cells acquire non-random orientations during cellular division. Wong et al.202 mentioned that the lipid phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) regulates actin movements and is present in the cleavage furrow. As cytokinesis progresses, this PtdIns(4,5)P2 is used, which is important for furrow durability. At the midbody, Rab35203 and 27 kDa inositol polyphosphate phosphatase-interacting protein A (IPIP27)204 coordinate the PtdIns(4,5)P2 phosphatase OCRL. Through hydrolysis of PtdIns(4,5)P2, OCRL allows midbody actin to be removed for effective abscission203. Phosphatidylinositol 4-phosphate (PtdIns(4)P), which is the product of PtdIns(4,5)P2 hydrolysis, can maintain the PtdIns(4)P-binding proteins required for cytokinesis205. As a result, a phosphoinositide transformation cascade might be used to modulate cytokinesis spatially and temporally. To understand the mechanism underlying this placement, more research will be required. The Rab35 interacts with recruits and activates the oxidoreductase MICAL1, which depolymerizes and decondenses actin filaments and facilitates actin cleanup206. Other members of the MICAL family help to orient vesicles within the midbody. The Rab11-positive endosomes are transported by MICAL-L1207 and Rab8-positive vesicles are tethered by MICAL3208. These suggest that this family of proteins has an important role in synchronizing cytokinetic consequences.
Membrane dynamics in small organelles: Cells also contain many smaller membrane-bound structures whose biological roles are still under research. The inheritances of these small organelles are less compactly regulated than that of huge single-copy organelles previously described as they are usually numerous and dynamic209. The membrane of pre-existing peroxisome lengthens to create a tubule during the mitotic division, which subsequently constricts and eventually undergoes scission to produce new peroxisomes210. Fate-determining endosomes are dynamically separated at the midzone microtubules with the support of kinesin motors in unevenly dividing sensory organs in D. melanogaster with asymmetric spindle disarrangement directing endosomal transmission211. Recent researches suggest that endocytic membrane transit may play a role in cell plate development. In the near future, one main area of study can be revealed by dealing with the mechanisms of symmetric versus asymmetric inheritance of endosomes212. During the time of mitotic exit, Endosomes are also needed213,214. Cytokinesis failure is the result of the shutting down of endosomal sorting due to the minute molecule prazosin215. Exocyst-dependent vesicular union has been proposed as a mechanism for total abscission in previous investigations216. However, more recent proof reports that endosomes are positive for Rab11 family-interconnecting protein 3 (Rab11-FIP3) or Rab35 convey proteins needed for the finishing of cytokinesis217-219. The prevalence of intra-midbody vesicles decreases as midbodies mature, indicating that the need for these union events decreases as cells attain abscission213. Near the future abscission site, both Rab11-FIP3-positive and Rab35-positive endosomes are stored to produce secondary ingression which is an intra-midbody construction that specifies the final cut site206,220. According to Pohl and Jentsch221, it must eliminate and/or reuse midbody remains of post-abscission at the end of cell division. This could be attributable to autophagy, although other evidence suggests that phagocytosis completes the process222. However, more research in the future is needed to confirm the actual roles of autophagy and the division of autophagosomes at the time of cell division. With a few exceptions, each cell in the G1 phase contains just one centrosome, which is made up of two centrioles and pericentriolar materials (PCM) and which need to be replicated before cell division occurs so that the two newly created cells both inherit a single centrosome223. During interphase, many organelles in plant cells have a more or less random distribution, but during mitosis and/or cytokinesis, they assume fairly definite positions224. The synthesis and turnover of lipid droplets, which serve as repositories for membrane extension and energy generation, are tightly regulated225. A recent report by Cruz et al.226 proposed that the cell cycle regulates the lipid droplet number and localization at the time of division. Peroxisomes can be replicated by division or can also be formed de novo from the ER. At the time of mitosis, these peroxisomes are oriented together around spindle poles to maintain the symmetric inheritance227,228. The deletion of the peroxisomal protein PEX11B causes uneven peroxisome transmission, mismatched mitotic spindles and aberrant division in epidermal progenitor cells229. Midbodies help in abscission without which cell division is incomplete and it needs the help of membrane and cytoskeletal reorganizing activities230.
Dynamics of plasma membranes: The plasma membrane of the cells at the time of division takes various curve-like structures as they react to the mechanical stress due to their close attachment to the cortex and their rearrangement that supports the enhancement of the surface area of the cells. According to Ramkumar and Baum231, cell volume, as well as surface topology, changes gradually at the time of mitosis. Mitosis begins with a dramatic series of changes in cell shape, motility and polarity for many animal cells232-234. Cells go through the shaping of the membrane, delivery and removal as there is very little time for de novo membrane synthesis. The membrane’s architecture and functional activity vary not just between specimens but also within cell populations235. Newly formed membranes emerge from internal parts through cell trafficking methods like exocytosis or endocytosis recycling236 or through plasma membrane restructuring237. During diffuse development, enzymes, carbohydrates and other elements of the cell wall are continuously and actively transferred to and from the plasma membrane. Vesicles carrying cell wall components travel along actin and microtubule-based cytoskeletal pathways. During cytokinesis, many of these elements as well as other proteins, vesicles and lipids are transported to and from the cell plate238. At the anaphase stage, the plasma membrane expands and this process helps the spindle to take a proper position for maintaining symmetry in the division. The architecture of the plasma membrane and the location of the spindle has an inverse relationship. The spindle position perturbation relocates the apparatus during the asymmetrical expansion of the cortex membrane through membrane blebbing237. The cleavage furrow location, which is controlled by the polar nature of membrane blebbing through rhythmic motion along the cortex, relieves membrane tension created by repeated changes in membrane shape throughout the division phase239. Expansion of the polar membrane is the cause of a driving force to help the furrow ingression in the embryos of sea urchins240. Different internal matters are acting as a helping hand in membrane expansion that is not yet known but all the above-mentioned examples prove that different cells have different pathways for membrane expansion. There is an intense relationship between the endocytic network and the plasma membrane. The plasma membrane serves as the entry point into the endocytic network. There are two different opinions about the endocytosis is continued or not during the time of cell division. Some proposed that there is no endocytosis at the time of cell division241 and according to others, endocytic recycling can be the means of plasma membrane expansion236. But the most followed opinion says that some amount of endocytosis is carried out during the division. Growth factor receptors and other important endocytosis cargoes have been given the first chance for their partitioning during division242.
Plasma membrane is connected to the cell cortex: The plasma membrane consists of some lipids and proteins that interact with the cortex in a highly structured, organized and constrained manner243-245. These physical limits are crucial in terms of functionality. Schmidt and Nichols246 reported that in the cleavage furrow, a diffusion barrier is present. It is attached to both the membranes and also with the cytoskeleton through the help of filament-forming septin proteins. Measurements of lipid and protein dispersion rates in remanufactured synthetic bilayers surpass cellular membranes with similar lipid content by more than a factor of ten247. Various additional membrane-binding adaptors help the plasma membrane of a dividing cell to make a connection with the cell cortex27. It includes anillin248, ezrin, radixin and moesin (ERM) proteins249. The Rho, a small GTPase and its deformers are important controllers of cytoskeletal mobility that work in conjunction with the plasma membrane249-252. Some other proteins that control the actin cytoskeleton have lipid-binding domains in them such as capping protein253, profilin254, cofilin255 and myosin II208. Actin filament polymerization against membranes generates force for several cellular activities, including organelle dynamics, endocytosis, phagocytosis, migration and morphogenesis256. A synchronized pattern of cell shape modification characterizes both cell division and cell migration. Despite their apparent functional differences, these two processes may have identical mechanisms and signaling pathways when it comes to cell shape257. These crucial linkers or responder proteins are not insider membrane proteins; instead, they are linked to the membrane by lipid alterations or lipid-attaching regions. The reproducing cell can control and perform membrane modifications with high spatial and temporal accuracy due to bidirectional interconnections between the membrane as well as the cortex. Throughout cytokinesis, daughter cells are irrevocably separated by furrow build-up and plasma membrane fission258. Knowing the processes underlying the regulated restructuring of the plasma membrane and its interaction with the cortex in cell cultures and 3D tissue settings will be a huge task in the coming years259.
Cytokinetic abscission: Our current understanding of how cytokinesis is completed has been fueled by the midbody location and functional requirements for ESCRT proteins in cytokinetic abscission10. Although the intercellular tract holding the midbody is one of the most visible structures in dividing animal cells, its role in the abscission phase of cytokinesis is mostly unclear260. As reported by Fabbro et al.261 a portion of the central pindling complex, MKLP1 involves the centrosomal protein CEP55 in the midbody. In turn CEP55, by direct connection with upstream ESCRT elements TSG101 and ALIX engages the ESCRT-III machinery262-266. During anaphase, cytokinesis in animal cells begins with the formation of a central spindle. Active ESCRT-III filament polymerization is believed to operate cytokinetic abscission213,266,267. By super-resolution approaches, it has recently become known that spiral filaments of ESCRT-III broaden the abscission area268. In various cell culture mechanisms and certain tissues, ESCRT-III is crucial for cytokinesis. For instance, it can be mentioned about the germline stem cells in D. melanogaster269,270, in mice271 and some crenarchaea146. This shows that as animals’ sophistication has grown, this machinery’s ancestral job in cell replication has been co-opted for additional membrane restructuring processes. It is recently shown by Karasmanis et al.266 that at the midbody bi-ringed membrane-bound septins direct orderly ESCRT assembly during cytokinesis. As a result, imaging-based experiments have been the primary research technique for uncovering many elements of the molecular signaling pathways that govern mitosis and cytokinesis160. In yeast cytokinesis, septins have important roles as connections between septins and controllers of cytokinesis in metazoans such as Rab35219 and GOLPH3205 were recognized.
| Table 1: | Biophysical techniques for studying the membrane dynamics | |||
| Techniques | Advantages | References |
| Confocal microscopy | For following membrane dynamics on a time scale (flat membranes can easily studied in the focused thin sections) |
Minsky282 |
| Fluorescence recovery after Photo-bleaching (FRAP) |
Gives data about the dynamic nature of the sample fluorophores |
Snapp et al.290 |
| Total internal reflection Fluorescence (TIRF) microscopy |
Detection of single molecules, high temporal resolution for observing the dynamic phenomena of the surface |
Groves et al.289 |
| Single particle tracking (SPT) | Very useful method for distinguishing different motion types of the particles |
García-Sáez and Schwille286 |
| Fluorescence correlation Spectroscopy (FCS) |
Powerful in characterization of the fluidity and the state of lipid bilayers |
Haustein and Schwille291 |
| Fluorescence interference Contrast microscopy (FLIC) |
More topographically quantifiable, greater range than TIRF | Groves et al.289 |
| Image correlation spectroscopy (ICS) |
Spatial correlation studies of the fluctuations in fluorescence intensity of an image |
Petersen et al.292 |
| Spatio-temporal image correlation spectroscopy (STICS) |
Whole spatial and temporal correlation function analysis of a stack of images is possible |
Hebert et al.293 |
| Particle image correlation spectroscopy (PICS) |
Combination of ICS and SPT | Semrau and Schmidt294 |
| Secondary ion mass spectroscopy | Gives us a map of the chemical constitution of supported bilayers with phase distinction |
Kraft et al.295 |
| Stimulated emission depletion microscopy (STED) |
Improve the resolution to a few 10 nm | Nägerl et al.296 |
| Photoactivated localization microscopy (PALM) |
Increases resolution | Sengupta et al.297 |
| Near-field scanning optical microscopy (NSOM) |
Estimates the fluorescence intensity on the surface of the membrane with a very sharp optical fiber |
Ianoul and Johnston298 |
Interactions between cells affect cell division: The extracellular environment and the nearby cells of the dividing cell have a deep connection during the division stage231,272. According to Roubinet et al.257 a force is required for all the changes that happen in mitotic rounding, ingression of the cleavage furrow and sealing after abscission273. If consider the monolayers of the epithelium at the time of mitotic entry, the tension between cells decreases, as well as the cytokinetic progress is put forward by the neighbouring cells. However, if the adhesive force is more than required then cytokinesis failure may happen274,275. All of these prove that every cell has a connection with the external environment as well as its nearby cells during division and there are also many mechanical signals and signaling cascades that are there for maintaining this connection with nearby cells. Changes to cellular mechanics are brought on by both exogenous (contact, wounding, dehydration, floods, pathogen invasion, gravity) and endogenous (expansion, cell mobility, division, morphogenesis) sources, which must be detected and responded to276. Plant development and its morphogenesis can be influenced by the cell cycle regulating and controlling mechanism. However, all the cell cycle regulation is more complicated than simply speeding up or lowering cell synthesis, which results in quicker or slower growth277. Depletion of cell plasma membrane inhabited proteins such as particular G protein-coupled sensors can inhibit cell division278,279. As proposed by Özlü et al.280 mitotic and interphase cell surface proteomes are different. This opens up new possibilities for signals that connect the cell division process to the cell’s external environment.
Techniques to study biological membrane dynamics: During the previous few years, understanding of the molecular structure of biological membranes has substantially improved due to the explosion of new technical interventions that have made it possible to examine membrane surfaces in unimaginable ways281. There are several novel methods for studying the membrane dynamics available most of them are microscopy-based (Table 1). Due to these recent advances, membrane and organelle dynamics during cell division can easily be examined. These techniques help us to examine the physical, biochemical as well as biophysical changes of the cellular membranes during the whole cell division process282. Confocal microscopy is the most widely used technique in many laboratories for the routine study of biological membranes. To maximize its spatial resolution power, it exclusively measures the fluorescence of thin sample slices282. In confocal microscopy, in front of the detector, a mini pinhole is kept for imaging only the fluorescence coming from the focus of the sample. A laser beam is used as a light source to facilitate the scanning of the sample. Fluorescence recovery after photo-bleaching (FRAP) is a technique or mechanism that is used in confocal microscopes to let us know about the dynamic nature of the fluorophores in the sample281. The foundation of FRAP is the study of the temporal rise in emission intensity in a sample area that has previously undergone photobleaching under intense laser illumination. But in this technique, high illumination power is required which often proves harmful to the sample and even to its dynamics283. Total Internal Reflection Fluorescence (TIRF) Microscopy uses the optically dense medium like glass and the less dense medium like an aqueous solution to its complete internal reflection to limit illumination to a range of 100-200 nm above the glass/water boundary284. In TIRF, data are assembled with cameras that have a huge resolution. The main advantage of this technique is it allows to detection of even the change of a single molecule284. In addition, single particle tracking (SPT) can be used to track the movement of individual lipids or proteins inside membranes247,285,286. In this technique, Nanoparticles coated with ligands (like antibodies) are used to label the samples. The SPT develops into a very potent technique for identifying different motion patterns such as directed flow, irregular diffusion, confined diffusion and Brownian diffusion287. Fluorescence correlation spectroscopy (FCS) is based on the statistical assessment of fluctuations in fluorescence over time, which are typically brought on by the diffusion of a single fluorophore into and out of the sensor container288. Characterizing the fluidity and state of lipid bilayers of various complexity has shown to be highly helpful286. In fluorescence interference contrast (FLIC) microscopy fluorescent probes are placed close to a reflective plane289. The fluorescence intensity is modulated and the collected data can be used to map topography at the nanoscale level. This has a greater range than TIRF as it allows more topographically quantifiable limits289.
Technological intervention to achieve conceptual advances: Understanding the dynamic regulation of organelles and sub-organellar structures at the time of division would be difficult until we can anticipate and regulate them in high resolution non-invasively. During mitosis, several membrane compartments undergo rigorous spatiotemporal reorganization. Nowadays, several new technologies such as lattice light sheet microscopy with adaptive optics have evolved for high-resolution live imaging of cellular organelles at the time of their partition134. The 4D imaging data have permitted to making of a dynamic protein atlas of cell division299. The cell is made up of DNA along with complex elements that must be meticulously passed on to the offspring. Several new technologies are being developed that allow us to see several new insights into the division process. Modern imaging devices and organelle-specific probes are being created. The capacity to precisely alter division structures utilizing novel approaches like optogenetics catalyzes technological intervention to study membrane dynamics300-302. Distinct intracellular membrane groups with different roles appear to have discovered different methods to ensure proper transmission21. Although symmetrically dividing cultured cells have been researched extensively, mechanical conditions in 3D and uneven situations may also be important for organelle heredity and appropriate division. In the future, the knowledge about these processes will surely increase with the evolution of revised models to enquire about the division in more physiologically relevant ways. Combining data from several areas of inquiry will be a major conceptual issue. This is challenging and tough to undertake since the primary viewpoints will certainly be achieved by exploring different fields and dimensions to gather atomic relationships that determine lipid behaviour to organelle migration in dividing cells303. Life propagation necessitates adequate knowledge to duplicate life to understand the knowledge of cell division. For instance, it is not well understood how the specification and molecular interactions of membrane proteins and lipids involved in organelle remodeling disrupt organellar function. Intracellular membrane sections tasked with various activities seem to have devised several strategies for ensuring proper transmission27. During mitosis, several membrane sections undergo extensive spatiotemporal reorganization. These precisely orchestrated organizational shifts are critical to completing the division cycle to guarantee viable offspring. The role of kinases in organellar dissociation and cellular rearrangements for mitotic entry is well understood, but how this phosphorylation is restored mechanistically is rarely documented in the journal. In upcoming years, a better knowledge of the regional roles of phosphatases will be a major challenge. The knowledge about the contribution and organization of splicing speckles, stress granules and centrioles on membrane-less organelles in cell biology at the time of division is very limited304. According to some current investigations, the protein kinase DYRK3 works as mitotic dissolves assisting in the solubilization of several of the above-mentioned structures. However, it is yet unknown why these structures must be disassembled during division305-307. Even though the knowledge of intracellular compartment inheritance is growing, our comprehension of the molecular pathways driving the equitable distribution of organelles among two daughter cells is still lacking to answer many critical and fascinating topics. It will be hard to define membrane and organelle movements in cell division without knowing how they associate with membrane-bound organelles that are similarly characterized by weak collective interactions within the membrane.
CONCLUSION AND FUTURE PROSPECTS
The cytoskeleton and different signaling cascades drive the cells carefully to strategize their genetic matter division and their physical partitioning. But still, there are enumerable questions that remain about this topic. The disassociation, separation and reassociation of ER, Golgi and nuclear envelope-like larger organelles are carefully controlled. But previously it was believed that mitochondria and an endo-lysosomal system like smaller organelles were partitioned stochastically. In recent studies, it is revealed that they are also partitioned actively. In the coming years, it will be extremely difficult to combine this knowledge to comprehend how the remodeling of various organelles is linked and how changes in organelle function and structure affect the cell division mechanism. Cells have developed complicated organelle inheritance systems to achieve productive intracellular organelle sharing. Below, we outline several significant areas and breakthroughs that will revolutionize our knowledge of this field and illuminate the theoretical advancements required to achieve these breakthroughs.
Understanding the kinetics of biological membranes during cell division is crucial for both plant and animal biologists. Membranes are crucial for preserving the integrity of cells and controlling molecular transport, both of which are necessary for cell division. Plant processes such as cytokinesis, in which the membrane's behaviour directs the development of a new cell wall, are influenced by membrane dynamics. Understanding membrane dynamics during cell division also provides insights into haploid induction in maize, where abnormality in membrane behavior can lead to the formation of haploid seeds. For animal cells to divide equally among their daughter cells during mitosis, membrane alterations are essential. Biologists can learn more about how aberrations in membrane behaviour may result in illnesses or defects in development by examining these kinetics. Furthermore, by using this knowledge, specific interventions may be developed to rectify these disturbances, which will ultimately increase our capacity to control cell division for a variety of purposes, including medical therapy and agricultural improvement.
SIGNIFICANCE STATEMENT
Since cell division is an essential process for the survival and reproduction of eukaryotic cells, this review is important because it offers a thorough synthesis of current information on the dynamics of biological membranes during this process. Reviewing the relationship between these components and their effects on cell division, the paper focuses on lipid composition, membrane compartmentalization and the processes behind organelle remodelling, distribution and inheritance. It also highlights how important cell-cycle proteins are in controlling and preserving the changes between mitosis, cytokinesis and interphase. Discovering the intricacies of cellular reproduction requires an understanding of these dynamics, which has ramifications for cell biology, genetics and related disciplines of study.
ACKNOWLEDGMENT
Author is thankful to the Indian Council of Agricultural Research (ICAR) for providing ICAR-NTS(PG) to pursue the master’s program. All the authors acknowledge reviewers for their valuable comments and suggestions.
REFERENCES
- Neumüller, R.A. and J.A. Knoblich, 2009. Dividing cellular asymmetry: Asymmetric cell division and its implications for stem cells and cancer. Genes Dev., 23: 2675-2699.
- Sclafani, R.A. and T.M. Holzen, 2007. Cell cycle regulation of DNA replication. Annu. Rev. Genet., 41: 237-280.
- Sablowski, R. and C. Gutierrez, 2022. Cycling in a crowd: Coordination of plant cell division, growth, and cell fate. Plant Cell, 34: 193-208.
- Nasmyth, K., 2002. Segregating sister genomes: The molecular biology of chromosome separation. Science, 297: 559-565.
- Walczak, C.E., S. Cai and A. Khodjakov, 2010. Mechanisms of chromosome behaviour during mitosis. Nat. Rev. Mol. Cell Biol., 11: 91-102.
- Musacchio, A., 2015. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol., 25: R1002-R1018.
- Daum, J.R., C.O. DuBose, S. Sivakumar and G.J. Gorbsky, 2022. Live Fluorescence Imaging of Chromosome Segregation in Cultured Cells. In: Mitosis: Methods and Protocols, Hinchcliffe, E.H. (Ed.), Humana, New York, ISBN: 978-1-0716-1904-9, pp: 61-86.
- Green, R.A., E. Paluch and K. Oegema, 2012. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol., 28: 29-58.
- Mierzwa, B. and D.W. Gerlich, 2014. Cytokinetic abscission: Molecular mechanisms and temporal control. Dev. Cell, 31: 525-538.
- Andrade, V. and A. Echard, 2022. Mechanics and regulation of cytokinetic abscission. Front. Cell Dev. Biol., 10.
- Nishi, M., K. Hu, J.M. Murray and D.S. Roos, 2008. Organellar dynamics during the cell cycle of Toxoplasma gondii. J. Cell Sci., 121: 1559-1568.
- Sheahan, M.B., R.J. Rose and D.W. McCurdy, 2004. Organelle inheritance in plant cell division: The actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J., 37: 379-390.
- Mishra, P. and D.C. Chan, 2014. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol., 15: 634-646.
- Echard, A., G.R.X. Hickson, E. Foley and P.H. O'Farrell, 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol., 14: 1685-1693.
- Eggert, U.S., A.A. Kiger, C. Richter, Z.E. Perlman, N. Perrimon, T.J. Mitchison and C.M. Field, 2004. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol., 2.
- Skop, A.R., H. Liu, J. Yates, B.J. Meyer and R. Heald, 2004. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science, 305: 61-66.
- Sönnichsen, B., L.B. Koski, A. Walsh, P. Marschall and B. Neumann et al., 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature, 434: 462-469.
- Kittler, R., L. Pelletier, A.K. Heninger, M. Slabicki and M. Theis et al., 2007. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat. Cell Biol., 9: 1401-1412.
- Neumann, B., T. Walter, J.K. Hériché, J. Bulkescher and H. Erfle et al., 2010. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature, 464: 721-727.
- Atilla-Gokcumen, G.E., A.B. Castoreno, S. Sasse and U.S. Eggert, 2010. Making the cut: The chemical biology of cytokinesis. ACS Chem. Biol., 5: 79-90.
- Jongsma, M.L.M., I. Berlin and J. Neefjes, 2015. On the move: Organelle dynamics during mitosis. Trends Cell Biol., 25: 112-124.
- Gould, G.W., 2016. Animal cell cytokinesis: The role of dynamic changes in the plasma membrane proteome and lipidome. Semin. Cell Dev. Biol., 53: 64-73.
- Champion, L., M.I. Linder and U. Kutay, 2017. Cellular reorganization during mitotic entry. Trends Cell Biol., 27: 26-41.
- Wang, B., H. Wang, Y. Li and L. Song, 2022. Lipid metabolism within the bone micro-environment is closely associated with bone metabolism in physiological and pathophysiological stages. Lipids Health Dis., 21.
- Cauvin, C. and A. Echard, 2015. Phosphoinositides: Lipids with informative heads and mastermind functions in cell division. Biochim. Biophys. Acta Mol. Cell Biol. Lipids, 1851: 832-843.
- Brindley, D.N., 2004. Lipid phosphate phosphatases and related proteins: Signaling functions in development, cell division, and cancer. J. Cell. Biochem., 92: 900-912.
- Storck, E.M., C. Özbalci and U.S. Eggert, 2018. Lipid cell biology: A focus on lipids in cell division. Annu. Rev. Biochem., 87: 839-869.
- Harayama, T. and H. Riezman, 2018. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol., 19: 281-296.
- van Meer, G., D.R. Voelker and G.W. Feigenson, 2008. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol., 9: 112-124.
- Sharpe, H.J., T.J. Stevens and S. Munro, 2010. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell, 142: 158-169.
- Scorrano, L., M.A. de Matteis, S. Emr, F. Giordano and G. Hajnóczky et al., 2019. Coming together to define membrane contact sites. Nat. Commun., 10.
- Muro, E., G.E. Atilla-Gokcumen and U.S. Eggert, 2014. Lipids in cell biology: How can we understand them better? Mol. Biol. Cell, 25: 1819-1823.
- Ng, M.M., F. Chang and D.R. Burgess, 2005. Movement of membrane domains and requirement of membrane signaling molecules for cytokinesis. Dev. Cell, 9: 781-790.
- Atilla-Gokcumen, G.E., E. Muro, J. Relat-Goberna, S. Sasse and A. Bedigian et al., 2014. Dividing cells regulate their lipid composition and localization. Cell, 156: 428-439.
- Kunduri, G., U. Acharya and J.K. Acharya, 2022. Lipid polarization during cytokinesis. Cells, 11.
- Sprong, H., P. van der Sluijs and G. van Meer, 2001. How proteins move lipids and lipids move proteins. Nat. Rev. Mol. Cell Biol., 2: 504-513.
- Ali, U., S. Lu, T. Fadlalla, S. Iqbal and H. Yue et al., 2022. The functions of phospholipases and their hydrolysis products in plant growth, development and stress responses. Prog. Lipid Res., 86.
- Kwok, A.C.M. and J.T.Y. Wong, 2005. Lipid biosynthesis and its coordination with cell cycle progression. Plant Cell Physiol., 46: 1973-1986.
- Atilla-Gokcumen, G.E. , A.V. Bedigian, S. Sasse and U.S. Eggert, 2011. Inhibition of glycosphingolipid biosynthesis induces cytokinesis failure. J. Am. Chem. Soc., 133: 10010-10013.
- Echard, A., 2012. Phosphoinositides and cytokinesis: The “PIP” of the iceberg. Cytoskeleton, 69: 893-912.
- Holy, J. and E. Perkins, 2009. Structure and Function of the Nucleus and Cell Organelles. In: Bioinformatics for Systems Biology, Krawetz, S. (Ed.), Humana Press, Totowa, New Jersey, ISBN: 978-1-59745-440-7, pp: 3-31.
- Zhao, Y.G. and H. Zhang, 2020. Phase separation in membrane biology: The interplay between membrane-bound organelles and membraneless condensates. Dev. Cell, 55: 30-44.
- Voeltz, G.K., M.M. Rolls and T.A. Rapoport, 2002. Structural organization of the endoplasmic reticulum. EMBO Rep., 3: 944-950.
- McBride, H.M., M. Neuspiel and S. Wasiak, 2006. Mitochondria: More than just a powerhouse. Curr. Biol., 16: R551-R560.
- Howe, C.J., A.C. Barbrook, R.E.R Nisbet, P.J. Lockhart and A.W.D. Larkum, 2008. The origin of plastids. Philos. Trans. R. Soc. B, 363: 2675-2685.
- Pfeffer, S., F. Brandt, T. Hrabe, S. Lang, M. Eibauer, R. Zimmermann and F. Förster, 2012. Structure and 3D arrangement of endoplasmic reticulum membrane-associated ribosomes. Structure, 20: 1508-1518.
- Guo, Y., D.W. Sirkis and R. Schekman, 2014. Protein sorting at the trans-Golgi network. Annu. Rev. Cell Dev. Biol., 30: 169-206.
- Doxsey, S., D. McCollum and W. Theurkauf, 2005. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol., 21: 411-434.
- Dell'Angelica, E.C., C. Mullins, S. Caplan and J.S. Bonifacino, 2000. Lysosome-related organelles. FASEB J., 14: 1265-1278.
- Islinger, M., A. Voelkl, H.D. Fahimi and M. Schrader, 2018. The peroxisome: An update on mysteries 2.0. Histochem. Cell Biol., 150: 443-471.
- Zheng, Z.L., 2022. Cyclin-dependent kinases and CTD phosphatases in cell cycle transcriptional control: Conservation across eukaryotic kingdoms and uniqueness to plants. Cells, 11.
- Imoto, Y., Y. Yoshida, F. Yagisawa, H. Kuroiwa and T. Kuroiwa, 2011. The cell cycle, including the mitotic cycle and organelle division cycles, as revealed by cytological observations. J. Electron Microsc., 60: S117-S136.
- Shimotohno, A., S.S. Aki, N. Takahashi and M. Umeda, 2021. Regulation of the plant cell cycle in response to hormones and the environment. Annu. Rev. Plant Biol., 72: 273-296.
- Nasa, I. and A.N. Kettenbach, 2018. Coordination of protein kinase and phosphoprotein phosphatase activities in mitosis. Front. Cell Dev. Biol., 6.
- Huguet, F., S. Flynn and P. Vagnarelli, 2019. The role of phosphatases in nuclear envelope disassembly and reassembly and their relevance to pathologies. Cells, 8.
- Kim, T., 2022. Recent progress on the localization of PLK1 to the kinetochore and its role in mitosis. Int. J. Mol. Sci., 23.
- Combes, G., I. Alharbi, L.G. Braga and S. Elowe, 2017. Playing polo during mitosis: PLK1 takes the lead. Oncogene, 36: 4819-4827.
- Fuller, B.G., M.A. Lampson, E.A. Foley, S. Rosasco-Nitcher and K.V. Le et al., 2008. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature, 453: 1132-1136.
- Afonso, O., I. Matos, A.J. Pereira, P. Aguiar, M.A. Lampson and H. Maiato, 2014. Feedback control of chromosome separation by a midzone Aurora B gradient. Science, 345: 332-336.
- Wang, E., E.R. Ballister and M.A. Lampson, 2011. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J. Cell Biol., 194: 539-549.
- Schmitz, M.H.A., M. Held, V. Janssens, J.R.A. Hutchins and O. Hudecz et al., 2010. Live-cell imaging RNAi screen identifies PP2A-B55α and importin-β1 as key mitotic exit regulators in human cells. Nat. Cell Biol., 12: 886-893.
- Gharbi-Ayachi, A., J.C. Labbé, A. Burgess, S. Vigneron and J.M. Strub et al., 2010. The substrate of greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science, 330: 1673-1677.
- Mochida, S., S.L. Maslen, M. Skehel and T. Hunt, 2010. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science, 330: 1670-1673.
- Cundell, M.J., R.N. Bastos, T. Zhang, J. Holder, U. Gruneberg, B. Novak and F.A. Barr, 2013. The BEG (PP2A-B55/ENSA/Greatwall) pathway ensures cytokinesis follows chromosome separation. Mol. Cell, 52: 393-405.
- Bollen, M., W. Peti, M.J. Ragusa and M. Beullens, 2010. The extended PP1 toolkit: Designed to create specificity. Trends Biochem. Sci., 35: 450-458.
- Vagnarelli, P., D.F. Hudson, S.A. Ribeiro, L. Trinkle-Mulcahy and J.M. Spence et al., 2006. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat. Cell Biol., 8: 1133-1142.
- Vagnarelli, P., S. Ribeiro, L. Sennels, L. Sanchez-Pulido and F. de Lima Alves et al., 2011. Repo-man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev. Cell, 21: 328-342.
- Wurzenberger, C., M. Held, M.A. Lampson, I. Poser, A.A. Hyman and D.W. Gerlich, 2012. Sds22 and Repo-Man stabilize chromosome segregation by counteracting Aurora B on anaphase kinetochores. J. Cell Biol., 198: 173-183.
- Qian, J., M. Beullens, B. Lesage and M. Bollen, 2013. Aurora B defines its own chromosomal targeting by opposing the recruitment of the phosphatase scaffold repo-man. Curr. Biol., 23: 1136-1143.
- Grallert, A., E. Boke, A. Hagting, B. Hodgson, Y. Connolly and J.R. Griffiths et al., 2015. A PP1-PP2A phosphatase relay controls mitotic progression. Nature, 517: 94-98.
- Bloom, J. and F.R. Cross, 2007. Multiple levels of cyclin specificity in cell-cycle control. Nat. Rev. Mol. Cell Biol., 8: 149-160.
- Uhlmann, F., F. Lottspeich and K. Nasmyth, 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature, 400: 37-42.
- Lindon, C. and J. Pines, 2004. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J. Cell Biol., 164: 233-241.
- Meghini, F., T. Martins, X. Tait, K. Fujimitsu, H. Yamano, D.M. Glover and Y. Kimata, 2016. Targeting of Fzr/Cdh1 for timely activation of the APC/C at the centrosome during mitotic exit. Nat. Commun., 7.
- Zylstra, A. and M. Heinemann, 2022. Metabolic dynamics during the cell cycle. Curr. Opin. Syst. Biol., 30.
- Borg, N.A. and V.M. Dixit, 2017. Ubiquitin in cell-cycle regulation and dysregulation in cancer. Annu. Rev. Cancer Biol., 1: 59-77.
- Sivakumar, S. and G.J. Gorbsky, 2015. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol., 16: 82-94.
- Horn, S.R., M.J. Thomenius, E.S. Johnson, C.D. Freel and J.Q. Wu et al., 2011. Regulation of mitochondrial morphology by APC/CCdh1-mediated control of Drp1 stability. Mol. Biol. Cell, 22: 1207-1216.
- Du, Y., S. Ferro-Novick and P. Novick, 2004. Dynamics and inheritance of the endoplasmic reticulum. J. Cell Sci., 117: 2871-2878.
- Mehrtash, A.B. and M. Hochstrasser, 2019. Ubiquitin-dependent protein degradation at the endoplasmic reticulum and nuclear envelope. Semin. Cell Dev. Biol., 93: 111-124.
- Güttinger, S., E. Laurell and U. Kutay, 2009. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol., 10: 178-191.
- de Leeuw, R., Y. Gruenbaum and O. Medalia, 2018. Nuclear lamins: Thin filaments with major functions. Trends Cell Biol., 28: 34-45.
- D’Angelo, M.A. and M.W. Hetzer, 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol., 18: 456-466.
- Chang, W., H.J. Worman and G.G. Gundersen, 2015. Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol., 208: 11-22.
- Boni, A., A.Z. Politi, P. Strnad, W. Xiang, M.J. Hossain and J. Ellenberg, 2015. Live imaging and modeling of inner nuclear membrane targeting reveals its molecular requirements in mammalian cells. J. Cell Biol., 209: 705-720.
- Ungricht, R., M. Klann, P. Horvath and U. Kutay, 2015. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J. Cell Biol., 209: 687-704.
- Graumann, K. and D.E. Evans, 2011. Nuclear envelope dynamics during plant cell division suggest common mechanisms between kingdoms. Biochem. J., 435: 661-667.
- Gomez-Cavazos, J.S. and M.W. Hetzer, 2012. Outfits for different occasions: Tissue-specific roles of nuclear envelope proteins. Curr. Opin. Cell Biol., 24: 775-783.
- Fernández-Jiménez, N. and M. Pradillo, 2020. The role of the nuclear envelope in the regulation of chromatin dynamics during cell division. J. Exp. Bot., 71: 5148-5159.
- Voeltz, G.K., W.A. Prinz, Y. Shibata, J.M. Rist and T.A. Rapoport, 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell, 124: 573-586.
- Hu, J., Y. Shibata, C. Voss, T. Shemesh and Z. Li et al., 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science, 319: 1247-1250.
- Nixon-Abell, J., C.J. Obara, A.V. Weigel, D. Li and W.R. Legant et al., 2016. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science, 354.
- Schroeder, L.K., A.E.S. Barentine, H. Merta, S. Schweighofer and Y. Zhang et al., 2019. Dynamic nanoscale morphology of the ER surveyed by STED microscopy. J. Cell Biol., 218: 83-96.
- Westrate, L.M., J.E. Lee, W.A. Prinz and G.K. Voeltz, 2015. Form follows function: The importance of endoplasmic reticulum shape. Annu. Rev. Biochem., 84: 791-811.
- Zhang, H. and J. Hu, 2016. Shaping the endoplasmic reticulum into a social network. Trends Cell Biol., 26: 934-943.
- Pradillo, M., D. Evans and K. Graumann, 2019. The nuclear envelope in higher plant mitosis and meiosis. Nucleus, 10: 55-66.
- Mall, M., T. Walter, M. Gorjánácz, I.F. Davidson, T.B.N. Ly-Hartig, J. Ellenberg and I.W. Mattaj, 2012. Mitotic lamin disassembly is triggered by lipid-mediated signaling. J. Cell Biol., 198: 981-990.
- Torvaldson, E., V. Kochin and J.E. Eriksson, 2015. Phosphorylation of lamins determine their structural properties and signaling functions. Nucleus, 6: 166-171.
- Bahmanyar, S., R. Biggs, A.L. Schuh, A. Desai and T. Müller-Reichert et al., 2014. Spatial control of phospholipid flux restricts endoplasmic reticulum sheet formation to allow nuclear envelope breakdown. Genes Dev., 28: 121-126.
- Audhya, A., A. Desai and K. Oegema, 2007. A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol., 178: 43-56.
- Beaudouin, J., D. Gerlich, N. Daigle, R. Eils and J. Ellenberg, 2002. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell, 108: 83-96.
- Salina, D., K. Bodoor, D.M. Eckley, T.A. Schroer, J.B. Rattner and B. Burke, 2002. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell, 108: 97-107.
- Hebbar, S., M.T. Mesngon, A.M. Guillotte, B. Desai, R. Ayala and D.S. Smith, 2008. Lis1 and Ndel1 influence the timing of nuclear envelope breakdown in neural stem cells. J. Cell Biol., 182: 1063-1071.
- Turgay, Y., L. Champion, C. Balazs, M. Held and A. Toso et al., 2014. SUN proteins facilitate the removal of membranes from chromatin during nuclear envelope breakdown. J. Cell Biol., 204: 1099-1109.
- Mori, M., K. Somogyi, H. Kondo, N. Monnier and H.J. Falk et al., 2014. An Arp2/3 nucleated F-actin shell fragments nuclear membranes at nuclear envelope breakdown in starfish oocytes. Curr. Biol., 24: 1421-1428.
- King, M.C., T.G. Drivas and G. Blobel, 2008. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell, 134: 427-438.
- Laurell, E., K. Beck, K. Krupina, G. Theerthagiri and B. Bodenmiller et al., 2011. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell, 144: 539-550.
- Yang, L., T. Guan and L. Gerace, 1997. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J. Cell Biol., 137: 1199-1210.
- Daigle, N., J. Beaudouin, L. Hartnell, G. Imreh, E. Hallberg, J. Lippincott-Schwartz and J. Ellenberg, 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol., 154: 71-84.
- de Castro, I.J., R.S. Gil, L. Ligammari, M.L.D. Giacinto and P. Vagnarelli, 2018. CDK1 and PLK1 coordinate the disassembly and reassembly of the nuclear envelope in vertebrate mitosis. Oncotarget, 9: 7763-7773.
- Linder, M.I., M. Köhler, P. Boersema, M. Weberruss and C. Wandke et al., 2017. Mitotic disassembly of nuclear pore complexes involves CDK1- and PLK1-mediated phosphorylation of key interconnecting nucleoporins. Dev. Cell, 43: 141-156.E7.
- Martino, L., S. Morchoisne-Bolhy, D.K. Cheerambathur, L. van Hove and J. Dumont et al., 2017. Channel nucleoporins recruit PLK-1 to nuclear pore complexes to direct nuclear envelope breakdown in C. elegans. Dev. Cell, 43: 157-171.E7.
- Lu, L., M.S. Ladinsky and T. Kirchhausen, 2009. Cisternal organization of the endoplasmic reticulum during mitosis. Mol. Biol. Cell, 20: 3471-3480.
- Lu, L., M.S. Ladinsky and T. Kirchhausen, 2011. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J. Cell Biol., 194: 425-440.
- Puhka, M., H. Vihinen, M. Joensuu and E. Jokitalo, 2007. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J. Cell Biol., 179: 895-909.
- Puhka, M., M. Joensuu, H. Vihinen, I. Belevich and E. Jokitalo, 2012. Progressive sheet-to-tubule transformation is a general mechanism for endoplasmic reticulum partitioning in dividing mammalian cells. Mol. Biol. Cell, 23: 2424-2432.
- Schweizer, N. and H. Maiato, 2015. Membrane-based mechanisms of mitotic spindle assembly. Commun. Integr. Biol., 8.
- Wang, S., H. Tukachinsky, F.B. Romano and T.A. Rapoport, 2016. Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. eLife, 5.
- Wang, S., R.E. Powers, V.A.M. Gold and T.A. Rapoport, 2018. The ER morphology-regulating lunapark protein induces the formation of stacked bilayer discs. Life Sci. Alliance, 1.
- Smyth, J.T., A.M. Beg, S. Wu, J.W. Putney and N.M. Rusan, 2012. Phosphoregulation of STIM1 leads to exclusion of the endoplasmic reticulum from the mitotic spindle. Curr. Biol., 22: 1487-1493.
- Schlaitz, A.L., J. Thompson, C.C.L. Wong, J.R. Yates and R. Heald, 2013. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev. Cell, 26: 315-323.
- Smyth, J.T., T.A. Schoborg, Z.J. Bergman, B. Riggs and N.M. Rusan, 2015. Proper symmetric and asymmetric endoplasmic reticulum partitioning requires astral microtubules. Open Biol., 5.
- Champion, L., S. Pawar, N. Luithle, R. Ungricht and U. Kutay, 2019. Dissociation of membrane-chromatin contacts is required for proper chromosome segregation in mitosis. Mol. Biol. Cell, 30: 427-440.
- Anderson, D.J. and M.W. Hetzer, 2007. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat. Cell Biol., 9: 1160-1166.
- Schellhaus, A.K., P. de Magistris and W. Antonin, 2016. Nuclear reformation at the end of mitosis. J. Mol. Biol., 428: 1962-1985.
- Anderson, D.J. and M.W. Hetzer, 2008. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J. Cell Biol., 182: 911-924.
- Samwer, M., M.W.G. Schneider, R. Hoefler, P.S. Schmalhorst, J.G. Jude, J. Zuber and D.W. Gerlich, 2017. DNA cross-bridging shapes a single nucleus from a set of mitotic chromosomes. Cell, 170: 956-972.E23.
- Allen, P.B., Y.G. Kwon, A.C. Nairn and P. Greengard, 1998. Isolation and characterization of PNUTS, a putative protein phosphatase 1 nuclear targeting subunit. J. Biol. Chem., 273: 4089-4095.
- Steen, R.L., S.B. Martins, K. Taskén and P. Collas, 2000. Recruitment of protein phosphatase 1 to the nuclear envelope by a-kinase anchoring protein Akap149 is a prerequisite for nuclear lamina assembly. J. Cell Biol., 150: 1251-1262.
- Booth, D.G., M. Takagi, L. Sanchez-Pulido, E. Petfalski and G. Vargiu et al., 2014. Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery. eLife, 3.
- Takagi, M., Y. Nishiyama, A. Taguchi and N. Imamoto, 2014. Ki67 antigen contributes to the timely accumulation of protein phosphatase 1γ on anaphase chromosomes. J. Biol. Chem., 289: 22877-22887.
- Karg, T., B. Warecki and W. Sullivan, 2015. Aurora B-mediated localized delays in nuclear envelope formation facilitate inclusion of late-segregating chromosome fragments. Mol. Biol. Cell, 26: 2227-2241.
- Taddei, A., F. Hediger, F.R. Neumann and S.M. Gasser, 2004. The function of nuclear architecture: A genetic approach. Annu. Rev. Genet., 38: 305-345.
- Liu, S., M. Kwon, M. Mannino, N. Yang, F. Renda, A. Khodjakov and D. Pellman, 2018. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature, 561: 551-555.
- van der Zanden, S,Y., M.L.M. Jongsma, A.C.M. Neefjes, I. Berlin and J. Neefjes, 2023. Maintaining soluble protein homeostasis between nuclear and cytoplasmic compartments across mitosis. Trends Cell Biol., 33: 18-29.
- Otsuka, S., A.M. Steyer, M. Schorb, J.K. Hériché and M.J. Hossain et al., 2018. Postmitotic nuclear pore assembly proceeds by radial dilation of small membrane openings. Nat. Struct. Mol. Biol., 25: 21-28.
- Otsuka, S., K.H. Bui, M. Schorb, M.J. Hossain and A.Z. Politi et al., 2016. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. eLife, 5.
- Clever, M., T. Funakoshi, Y. Mimura, M. Takagi and N. Imamoto, 2012. The nucleoporin ELYS/Mel28 regulates nuclear envelope subdomain formation in HeLa cells. Nucleus, 3: 187-199.
- Olmos, Y., L. Hodgson, J. Mantell, P. Verkade and J.G. Carlton, 2015. ESCRT-III controls nuclear envelope reformation. Nature, 522: 236-239.
- Vietri, M., K.O. Schink, C. Campsteijn, C.S. Wegner and S.W. Schultz et al., 2015. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature, 522: 231-235.
- Olmos, Y., A. Perdrix-Rosell and J.G. Carlton, 2016. Membrane binding by CHMP7 coordinates ESCRT-III-dependent nuclear envelope reformation. Curr. Biol., 26: 2635-2641.
- Gu, M., D. LaJoie, O.S. Chen, A. von Appen and M.S. Ladinsky et al., 2017. LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. Proc. Natl. Acad. Sci. U.S.A., 114: E2166-E2175.
- Campsteijn, C., M. Vietri and H. Stenmark, 2016. Novel ESCRT functions in cell biology: Spiraling out of control? Curr. Opin. Cell Biol., 41: 1-8.
- Schöneberg, J., I.H. Lee, J.H. Iwasa and J.H. Hurley, 2017. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol., 18: 5-17.
- Gatta, A.T. and J.G. Carlton, 2019. The ESCRT-machinery: Closing holes and expanding roles. Curr. Opin. Cell Biol., 59: 121-132.
- Samson, R.Y., T. Obita, S.M. Freund, R.L. Williams and S.D. Bell, 2008. A role for the ESCRT system in cell division in archaea. Science, 322: 1710-1713.
- Logan, D.C., 2006. Plant mitochondrial dynamics. Biochim. Biophys. Acta Mol. Cell Res., 1763: 430-441.
- Labbé, K., A. Murley and J. Nunnari, 2014. Determinants and functions of mitochondrial behavior. Annu. Rev. Cell Dev. Biol., 30: 357-391.
- Pangou, E. and I. Sumara, 2021. The multifaceted regulation of mitochondrial dynamics during mitosis. Front. Cell Dev. Biol., 9.
- Fenton, A.R., T.A. Jongens and E.L.F. Holzbaur, 2021. Mitochondrial dynamics: Shaping and remodeling an organelle network. Curr. Opin. Cell Biol., 68: 28-36.
- Kashatus, D.F., K.H. Lim, D.C. Brady, N.L.K. Pershing, A.D. Cox and C.M. Counter, 2011. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol., 13: 1108-1115.
- Kanfer, G. and B. Kornmann, 2016. Dynamics of the mitochondrial network during mitosis. Biochem. Soc. Trans., 44: 510-516.
- Park, Y.Y. and H. Cho, 2012. Mitofusin 1 is degraded at G2/M phase through ubiquitylation by MARCH5. Cell Div., 7.
- Rhoads, D.M. and C.C. Subbaiah, 2007. Mitochondrial retrograde regulation in plants. Mitochondrion, 7: 177-194.
- Adachi, Y., K. Itoh, T. Yamada, K.L. Cerveny and T.L. Suzuki et al., 2016. Coincident phosphatidic acid interaction restrains Drp1 in mitochondrial division. Mol. Cell, 63: 1034-1043.
- Katajisto, P., J. Döhla, C.L. Chaffer, N. Pentinmikko and N. Marjanovic et al., 2015. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science, 348: 340-343.
- Jajoo, R., Y. Jung, D. Huh, M.P. Viana, S.M. Rafelski, M. Springer and J. Paulsson, 2016. Accurate concentration control of mitochondria and nucleoids. Science, 351: 169-172.
- Lawrence, E.J., E. Boucher and C.A. Mandato, 2016. Mitochondria-cytoskeleton associations in mammalian cytokinesis. Cell Div., 11.
- López-Doménech, G., C. Covill-Cooke, D. Ivankovic, E.F. Halff and D.F. Sheehan et al., 2018. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J., 37: 321-336.
- Morgan, D.O., 2007. The Cell Cycle: Principles of Control. New Science Press, London, United Kingdom, ISBN-13: 9780878935086, Pages: 297.
- Malumbres, M. and M. Barbacid, 2009. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer, 9: 153-166.
- Álvarez‐Fernández, M. and M. Malumbres, 2014. Preparing a cell for nuclear envelope breakdown: Spatio-temporal control of phosphorylation during mitotic entry. BioEssays, 36: 757-765.
- Tábara, L.C., J.L. Morris and J. Prudent, 2021. The complex dance of organelles during mitochondrial division. Trends Cell Biol., 31: 241-253.
- Salazar-Roa, M. and M. Malumbres, 2017. Fueling the cell division cycle. Trends Cell Biol., 27: 69-81.
- Rabouille, C. and E. Jokitalo, 2003. Golgi apparatus partitioning during cell division (Review). Mol. Membr. Biol., 20: 117-127.
- Tachikawa, M., 2023. Theoretical approaches for understanding the self-organized formation of the Golgi apparatus. Dev. Growth Differ., 65: 161-166.
- Barr, F.A., M. Puype, J. Vandekerckhove and G. Warren, 1997. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell, 91: 253-262.
- Shorter, J., R. Watson, M.E. Giannakou, M. Clarke, G. Warren and F.A. Barr, 1999. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J., 18: 4949-4960.
- Diao, A., L. Frost, Y. Morohashi and M. Lowe, 2008. Coordination of golgin tethering and SNARE assembly: GM130 binds syntaxin 5 in a p115-regulated manner. J. Biol. Chem., 283: 6957-6967.
- Witkos, T.M. and M. Lowe, 2015. The Golgin family of coiled-coil tethering proteins. Front. Cell Dev. Biol., 3.
- Carcedo, C.H., M. Bonazzi, S. Spanò, G. Turacchio, A. Colanzi, A. Luini and D. Corda, 2004. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science, 305: 93-96.
- Colanzi, A., C.H. Carcedo, A. Persico, C. Cericola and G. Turacchio et al., 2007. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. EMBO J., 26: 2465-2476.
- Lin, C.Y., M.L. Madsen, F.R. Yarm, Y.J. Jang, X. Liu and R.L. Erikson, 2000. Peripheral Golgi protein GRASP65 is a target of mitotic polo-like kinase (Plk) and Cdc2. Proc. Natl. Acad. Sci. U.S.A., 97: 12589-12594.
- Sütterlin, C., P. Hsu, A. Mallabiabarrena and V. Malhotra, 2002. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell, 109: 359-369.
- Wang, Y., J. Seemann, M. Pypaert, J. Shorter and G. Warren, 2003. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J., 22: 3279-3290.
- Preisinger, C., R. Körner, M. Wind, W.D. Lehmann, R. Kopajtich and F.A. Barr, 2005. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J., 24: 753-765.
- Tang, D., H. Yuan, O. Vielemeyer, F. Perez and Y. Wang, 2012. Sequential phosphorylation of GRASP65 during mitotic Golgi disassembly. Biol. Open, 1: 1204-1214.
- Mascanzoni, F., R. Iannitti and A. Colanzi, 2022. Functional coordination among the Golgi complex, the centrosome and the microtubule cytoskeleton during the cell cycle. Cells, 11.
- Ayala, I., R. Crispino and A. Colanzi, 2019. GRASP65 controls Golgi position and structure during G2/M transition by regulating the stability of microtubules. Traffic, 20: 785-802.
- Persico, A., R.I. Cervigni, M.L. Barretta, D. Corda and A. Colanzi, 2010. Golgi partitioning controls mitotic entry through aurora-A kinase. Mol. Biol. Cell, 21: 3708-3721.
- Guizzunti, G. and J. Seemann, 2016. Mitotic Golgi disassembly is required for bipolar spindle formation and mitotic progression. Proc. Natl. Acad. Sci. U.S.A., 113: E6590-E6599.
- Levine, T.P., C. Rabouille, R.H. Kieckbusch and G. Warren, 1996. Binding of the vesicle docking protein p115 to Golgi membranes is inhibited under mitotic conditions. J. Biol. Chem., 271: 17304-17311.
- Nakamura, N., M. Lowe, T.P. Levine, C. Rabouille and G. Warren, 1997. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell, 89: 445-455.
- Lowe, M., C. Rabouille, N. Nakamura, R. Watson and M. Jackman et al., 1998. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell, 94: 783-793.
- Tang, D., Y. Xiang, S. de Renzis, J. Rink, G. Zheng, M. Zerial and Y. Wang, 2011. The ubiquitin ligase HACE1 regulates Golgi membrane dynamics during the cell cycle. Nat. Commun., 2.
- Huang, S., D. Tang and Y. Wang, 2016. Monoubiquitination of syntaxin 5 regulates Golgi membrane dynamics during the cell cycle. Dev. Cell, 38: 73-85.
- Zhang, X., H. Zhang and Y. Wang, 2014. Phosphorylation regulates VCIP135 function in Golgi membrane fusion during the cell cycle. J. Cell Sci., 127: 172-181.
- Zhang, X. and Y. Wang, 2015. Cell cycle regulation of VCIP135 deubiquitinase activity and function in p97/p47-mediated Golgi reassembly. Mol. Biol. Cell, 26: 2242-2251.
- Uchiyama, K., E. Jokitalo, M. Lindman, M. Jackman and F. Kano et al., 2003. The localization and phosphorylation of p47 are important for Golgi disassembly-assembly during the cell cycle. J. Cell Biol., 161: 1067-1079.
- Wei, J.H., Z.C. Zhang, R.M. Wynn and J. Seemann, 2015. GM130 regulates Golgi-derived spindle assembly by activating TPX2 and capturing microtubules. Cell, 162: 287-299.
- Altan-Bonnet, N., R.D. Phair, R.S. Polishchuk, R. Weigert and J. Lippincott-Schwartz, 2003. A role for Arf1 in mitotic Golgi disassembly, chromosome segregation, and cytokinesis. Proc. Natl. Acad. Sci. U.S.A., 100: 13314-13319.
- Altan-Bonnet, N., R. Sougrat and J. Lippincott-Schwartz, 2004. Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol., 16: 364-372.
- Rabouille, C., T. Misteli, R. Watson and G. Warren, 1995. Reassembly of Golgi stacks from mitotic Golgi fragments in a cell-free system. J. Cell Biol., 129: 605-618.
- Gao, J., A. Gao, H. Zhou and L. Chen, 2022. The role of metal ions in the Golgi apparatus. Cell Biol. Int., 46: 1309-1319.
- Lowe, M., N.K. Gonatas and G. Warren, 2000. The mitotic phosphorylation cycle of the Cis-Golgi matrix protein Gm130. J. Cell Biol., 149: 341-356.
- Tang, D., K. Mar, G. Warren and Y. Wang, 2008. Molecular mechanism of mitotic Golgi disassembly and reassembly revealed by a defined reconstitution assay. J. Biol. Chem., 283: 6085-6094.
- Tang, D., H. Yuan and Y. Wang, 2010. The role of GRASP65 in Golgi cisternal stacking and cell cycle progression. Traffic, 11: 827-842.
- Miller, P.M., A.W. Folkmann, A.R.R. Maia, N. Efimova, A. Efimov and I. Kaverina, 2009. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat. Cell Biol., 11: 1069-1080.
- Gaietta, G.M., B.N.G. Giepmans, T.J. Deerinck, W.B. Smith and L. Ngan et al., 2006. Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proc. Natl. Acad. Sci. U.S.A., 103: 17777-17782.
- Goss, J.W. and D.K. Toomre, 2008. Both daughter cells traffic and exocytose membrane at the cleavage furrow during mammalian cytokinesis. J. Cell Biol., 181: 1047-1054.
- Magliozzi, R., Z.I. Carrero, T.Y. Low, L. Yuniati and C. Valdes-Quezada et al., 2018. Inheritance of the Golgi apparatus and cytokinesis are controlled by degradation of GBF1. Cell Rep., 23: 3381-3391.E4.
- Wong, R., I. Hadjiyanni, H.C. Wei, G. Polevoy, R. McBride, K.P. Sem and J.A. Brill, 2005. PIP2 hydrolysis and calcium release are required for cytokinesis in Drosophila spermatocytes. Curr. Biol., 15: 1401-1406.
- Dambournet, D., M. Machicoane, L. Chesneau, M. Sachse and M. Rocancourt et al., 2011. Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat. Cell Biol., 13: 981-988.
- Carim, S.C., K.B. El Kadhi, G. Yan, S.T. Sweeney, G.R. Hickson, S. Carréno and M. Lowe, 2019. IPIP27 coordinates PtdIns(4,5)P2 homeostasis for successful cytokinesis. Curr. Biol., 29: 775-789.e7.
- Sechi, S., G. Colotti, G. Belloni, V. Mattei and A. Frappaolo et al., 2014. GOLPH3 is essential for contractile ring formation and Rab11 localization to the cleavage site during cytokinesis in Drosophila melanogaster. PLoS Genet., 10.
- Frémont, S., H. Hammich, J. Bai, H. Wioland and K. Klinkert et al., 2017. Oxidation of F-actin controls the terminal steps of cytokinesis. Nat. Commun., 8.
- Reinecke, J.B., D. Katafiasz, N. Naslavsky and S. Caplan, 2015. Novel functions for the endocytic regulatory proteins MICAL-L1 and EHD1 in mitosis. Traffic, 16: 48-67.
- Liu, X., S. Shu, N. Billington, C.D. Williamson and S. Yu et al., 2016. Mammalian nonmuscle myosin II binds to anionic phospholipids with concomitant dissociation of the regulatory light chain. J. Biol. Chem., 291: 24828-24837.
- Robinson, D.G., M.C. Herranz, J. Bubeck, R. Pepperkok and C. Ritzenthaler, 2007. Membrane dynamics in the early secretory pathway. Crit. Rev. Plant Sci., 26: 199-225.
- Carmichael, R.E. and M. Schrader, 2022. Determinants of peroxisome membrane dynamics. Front. Physiol., 13.
- Derivery, E., C. Seum, A. Daeden, S. Loubéry, L. Holtzer, F. Jülicher and M. Gonzalez-Gaitan, 2015. Polarized endosome dynamics by spindle asymmetry during asymmetric cell division. Nature, 528: 280-285.
- Sunchu, B. and C. Cabernard, 2020. Principles and mechanisms of asymmetric cell division. Development, 147.
- Guizetti, J., L. Schermelleh, J. Mäntler, S. Maar and I. Poser et al., 2011. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science, 331: 1616-1620.
- König, J., E.B. Frankel, A. Audhya and T. Müller-Reichert, 2017. Membrane remodeling during embryonic abscission in Caenorhabditis elegans. J. Cell Biol., 216: 1277-1286.
- Zhang, X., W. Wang, A.V. Bedigian, M.L. Coughlin, T.J. Mitchison and U.S. Eggert, 2012. Dopamine receptor D3 regulates endocytic sorting by a Prazosin-sensitive interaction with the coatomer COPI. Proc. Natl. Acad. Sci. U.S.A., 109: 12485-12490.
- Gromley, A., C. Yeaman, J. Rosa, S. Redick and C.T. Chen et al., 2005. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell, 123: 75-87.
- Fielding, A.B., E. Schonteich, J. Matheson, G. Wilson and X. Yu et al., 2005. Rab11-FIP3 and FIP4 interact with Arf6 and the Exocyst to control membrane traffic in cytokinesis. EMBO J., 24: 3389-3399.
- Wilson, G.M., A.B. Fielding, G.C. Simon, X. Yu and P.D. Andrews et al., 2005. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol. Biol. Cell, 16: 849-860.
- Kouranti, I., M. Sachse, N. Arouche, B. Goud and A. Echard, 2006. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr. Biol., 16: 1719-1725.
- Schiel, J.A., G.C. Simon, C. Zaharris, J. Weisz, D. Castle, C.C. Wu and R. Prekeris, 2012. FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nat. Cell Biol., 14: 1068-1078.
- Pohl, C. and S. Jentsch, 2009. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat. Cell Biol., 11: 65-70.
- Crowell, E.F., A.L. Gaffuri, B. Gayraud-Morel, S. Tajbakhsh and A. Echard, 2014. Engulfment of the midbody remnant after cytokinesis in mammalian cells. J. Cell Sci., 127: 3840-3851.
- Prigent, C. and R. Uzbekov, 2022. Duplication and segregation of centrosomes during cell division. Cells, 11.
- Nebenführ, A., 2007. Organelle Dynamics During Cell Division. In: Cell Division Control in Plants, Verma, D.P.S. and Z. Hong (Eds.), Springer, Berlin, Heidelberg, ISBN: 978-3-540-73487-1, pp: 195-206.
- Walther, T.C., J. Chung and R.V. Farese, 2017. Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol., 33: 491-510.
- Cruz, A.L.S., N. Carrossini, L.K. Teixeira, L.F. Ribeiro-Pinto, P.T. Bozza and J.P.B. Viola, 2019. Cell cycle progression regulates biogenesis and cellular localization of lipid droplets. Mol. Cell. Biol., 39.
- Kredel, S., F. Oswald, K. Nienhaus, K. Deuschle and C. Röcker et al., 2009. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS ONE, 4.
- Knoblach, B. and R.A. Rachubinski, 2016. How peroxisomes partition between cells. A story of yeast, mammals and filamentous fungi. Curr. Opin. Cell Biol., 41: 73-80.
- Asare, A., J. Levorse and E. Fuchs, 2017. Coupling organelle inheritance with mitosis to balance growth and differentiation. Science, 355.
- Frémont, S. and A. Echard, 2018. Membrane traffic in the late steps of cytokinesis. Curr. Biol., 28: R458-R470.
- Ramkumar, N. and B. Baum, 2016. Coupling changes in cell shape to chromosome segregation. Nat. Rev. Mol. Cell Biol., 17: 511-521.
- Matthews, H.K., U. Delabre, J.L. Rohn, J. Guck, P. Kunda and B. Baum, 2012. Changes in Ect2 localization couple actomyosin-dependent cell shape changes to mitotic progression. Dev. Cell, 23: 371-383.
- Rosa, A., E. Vlassaks, F. Pichaud and B. Baum, 2015. Ect2/Pbl acts via rho and polarity proteins to direct the assembly of an isotropic actomyosin cortex upon mitotic entry. Dev. Cell, 32: 604-616.
- Strzyz, P.J., H.O. Lee, J. Sidhaye, I.P. Weber, L.C. Leung and C. Norden, 2015. Interkinetic nuclear migration is centrosome independent and ensures apical cell division to maintain tissue integrity. Dev. Cell, 32: 203-219.
- Snijder, B., R. Sacher, P. Rämö, E.M. Damm, P. Liberali and L. Pelkmans, 2009. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature, 461: 520-523.
- Boucrot, E. and T. Kirchhausen, 2007. Endosomal recycling controls plasma membrane area during mitosis. Proc. Natl. Acad. Sci. U.S.A., 104: 7939-7944.
- Kiyomitsu, T. and I.M. Cheeseman, 2013. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell, 154: 391-402.
- Gu, Y. and C.G. Rasmussen, 2022. Cell biology of primary cell wall synthesis in plants. Plant Cell, 34: 103-128.
- Sedzinski, J., M. Biro, A. Oswald, J.Y. Tinevez, G. Salbreux and E. Paluch, 2011. Polar actomyosin contractility destabilizes the position of the cytokinetic furrow. Nature, 476: 462-466.
- Gudejko, H.F.M., L.M. Alford and D.R. Burgess, 2012. Polar expansion during cytokinesis. Cytoskeleton, 69: 1000-1009.
- Fielding, A.B., A.K. Willox, E. Okeke and S.J. Royle, 2012. Clathrin-mediated endocytosis is inhibited during mitosis. Proc. Natl. Acad. Sci. U.S.A., 109: 6572-6577.
- Hinze, C. and E. Boucrot, 2018. Endocytosis in proliferating, quiescent and terminally differentiated cells. J. Cell Sci., 131.
- Kusumi, A., T.K. Fujiwara, R. Chadda, M. Xie and T.A. Tsunoyama et al., 2012. Dynamic organizing principles of the plasma membrane that regulate signal transduction: Commemorating the fortieth anniversary of singer and Nicolson's fluid-mosaic model. Annu. Rev. Cell Dev. Biol., 28: 215-250.
- Trimble, W.S. and S. Grinstein, 2015. Barriers to the free diffusion of proteins and lipids in the plasma membrane. J. Cell Biol., 208: 259-271.
- Köster, D.V. and S. Mayor, 2016. Cortical actin and the plasma membrane: Inextricably intertwined. Curr. Opin. Cell Biol., 38: 81-89.
- Schmidt, K. and B.J. Nichols, 2004. A barrier to lateral diffusion in the cleavage furrow of dividing mammalian cells. Curr. Biol., 14: 1002-1006.
- Kusumi, A., C. Nakada, K. Ritchie, K. Murase and K. Suzuki et al., 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: High-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct., 34: 351-378.
- Piekny, A.J. and M. Glotzer, 2008. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr. Biol., 18: 30-36.
- Kunda, P., A.E. Pelling, T. Liu and B. Baum, 2008. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr. Biol., 18: 91-101.
- Jordan, S.N. and J.C. Canman, 2012. Rho GTPases in animal cell cytokinesis: An occupation by the one percent. Cytoskeleton, 69: 919-930.
- Lekomtsev, S., K.C. Su, V.E. Pye, K. Blight and S. Sundaramoorthy et al., 2012. Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature, 492: 276-279.
- Kotýnková, K., K.C. Su, S.C. West and M. Petronczki, 2016. Plasma membrane association but not midzone recruitment of RhoGEF ECT2 Is essential for cytokinesis. Cell Rep., 17: 2672-2686.
- Terry, S.J., F. Donà, P. Osenberg, J.G. Carlton and U.S. Eggert, 2018. Capping protein regulates actin dynamics during cytokinetic midbody maturation. Proc. Natl. Acad. Sci. U.S.A., 115: 2138-2143.
- Giansanti, M.G., S. Bonaccorsi, B. Williams, E.V. Williams, C. Santolamazza, M.L. Goldberg and M. Gatti, 1998. Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev., 12: 396-410.
- Kusano, K.I., H. Abe, T. Obinata and T. Obinata, 1999. Detection of a sequence involved in actin-binding and phosphoinositide-binding in the N-terminal side of cofilin. Mol. Cell. Biochem., 190: 133-141.
- Lappalainen, P., T. Kotila, A. Jégou and G. Romet-Lemonne, 2022. Biochemical and mechanical regulation of actin dynamics. Nat. Rev. Mol. Cell Biol., 23: 836-852.
- Roubinet, C., P.T. Tran and M. Piel, 2012. Common mechanisms regulating cell cortex properties during cell division and cell migration. Cytoskeleton, 69: 957-972.
- Dobbelaere, J. and Y. Barral, 2004. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science, 305: 393-396.
- Levental, I. and S.L. Veatch, 2016. The continuing mystery of lipid rafts. J. Mol. Biol., 428: 4749-4764.
- Steigemann, P. and D.W. Gerlich, 2009. Cytokinetic abscission: Cellular dynamics at the midbody. Trends Cell Biol., 19: 606-616.
- Fabbro, M., B.B. Zhou, M. Takahashi, B. Sarcevic and P. Lal et al., 2005. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev. Cell, 9: 477-488.
- Carlton, J.G. and J. Martin-Serrano, 2007. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science, 316: 1908-1912.
- Morita, E., V. Sandrin, H.Y. Chung, S.G. Morham, S.P. Gygi, C.K. Rodesch and W.I. Sundquist, 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J., 26: 4215-4227.
- Carlton, J.G., M. Agromayor and J. Martin-Serrano, 2008. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. U.S.A., 105: 10541-10546.
- Christ, L., E.M. Wenzel, K. Liestøl, C. Raiborg, C. Campsteijn and H. Stenmark, 2016. ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J. Cell Biol., 212: 499-513.
- Karasmanis, E.P., D. Hwang, K. Nakos, J.R. Bowen, D. Angelis and E.T. Spiliotis, 2019. A septin double ring controls the spatiotemporal organization of the ESCRT machinery in cytokinetic abscission. Curr. Biol., 29: 2174-2182.E7.
- Mierzwa, B.E., N. Chiaruttini, L. Redondo-Morata, J.M. von Filseck and J. König et al., 2017. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat. Cell Biol., 19: 787-798.
- Goliand, I., S. Adar-Levor, I. Segal, D. Nachmias, T. Dadosh, M.M. Kozlov and N. Elia, 2018. Resolving ESCRT-III spirals at the intercellular bridge of dividing cells using 3D STORM. Cell Rep., 24: 1756-1764.
- Eikenes, Å.H., L. Malerød, A.L. Christensen, C.B. Steen and J. Mathieu et al., 2015. ALIX and ESCRT-III coordinately control cytokinetic abscission during germline stem cell division in vivo. PLoS Genet., 11.
- Matias, N.R., J. Mathieu and J.R. Huynh, 2015. Abscission is regulated by the ESCRT-III protein shrub in Drosophila germline stem cells. PLoS Genet., 11.
- Iwamori, T., N. Iwamori, L. Ma, M.A. Edson, M.P. Greenbaum and M.M. Matzuk, 2010. TEX14 interacts with CEP55 to block cell abscission. Mol. Cell. Biol., 30: 2280-2292.
- Dix, C.L., H.K. Matthews, M. Uroz, S. McLaren and L. Wolf et al., 2018. The role of mitotic cell-substrate adhesion re-modeling in animal cell division. Dev. Cell, 45: 132-145.E3.
- Lafaurie-Janvore, J., P. Maiuri, I. Wang, M. Pinot and J.B. Manneville et al., 2013. ESCRT-III assembly and cytokinetic abscission are induced by tension release in the intercellular bridge. Science, 339: 1625-1629.
- Uroz, M., S. Wistorf, X. Serra-Picamal, V. Conte and M. Sales-Pardo et al., 2018. Regulation of cell cycle progression by cell-cell and cell-matrix forces. Nat. Cell Biol., 20: 646-654.
- Uroz, M., A. Garcia-Puig, I. Tekeli, A. Elosegui-Artola and J.F. Abenza et al., 2019. Traction forces at the cytokinetic ring regulate cell division and polyploidy in the migrating zebrafish epicardium. Nat. Mater., 18: 1015-1023.
- Codjoe, J.M., K. Miller and E.S. Haswell, 2022. Plant cell mechanobiology: Greater than the sum of its parts. Plant Cell, 34: 129-145.
- Beemster, G.T.S., F. Fiorani and D. Inzé, 2003. Cell cycle: The key to plant growth control? Trends Plant Sci., 8: 154-158.
- Zhang, X., A.V. Bedigian, W. Wang and U.S. Eggert, 2012. G protein-coupled receptors participate in cytokinesis. Cytoskeleton, 69: 810-818.
- Zhang, X. and U.S. Eggert, 2013. Non-traditional roles of G protein-coupled receptors in basic cell biology. Mol. BioSyst., 9: 586-595.
- Özlü, N., M.H. Qureshi, Y. Toyoda, B.Y. Renard and G. Mollaoglu et al., 2014. Quantitative comparison of a human cancer cell surface proteome between interphase and mitosis. EMBO J., 34: 251-265.
- Sankaran, J. and T. Wohland, 2020. Fluorescence strategies for mapping cell membrane dynamics and structures. APL Bioeng., 4.
- Minsky, M., 1988. Memoir on inventing the confocal scanning microscope. Scanning, 10: 128-138.
- Kovaleski, J.M. and M.J. Wirth, 1997. Peer reviewed: Applications of fluorescence recovery after photobleaching. Anal. Chem., 69: 600A-605A.
- Axelrod, D., 2001. Total internal reflection fluorescence microscopy in cell biology. Traffic, 2: 764-774.
- Lenne, P.F., L. Wawrezinieck, F. Conchonaud, O. Wurtz and A. Boned et al., 2006. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J., 25: 3245-3256.
- García-Sáez, A.J. and P. Schwille, 2007. Single molecule techniques for the study of membrane proteins. Appl. Microbiol. Biotechnol., 76: 257-266.
- Saxton, M.J. and K. Jacobson, 1997. Single-particle tracking: Applications to membrane dynamics. Annu. Rev. Biophys., 26: 373-399.
- Thompson, N.L., 2002. Fluorescence Correlation Spectroscopy. In: Techniques, Lakowicz, J.R. (Ed.), Springer, Boston, Massachusetts, ISBN: 978-0-306-47057-8, pp: 337-378.
- Groves, J.T., R. Parthasarathy and M.B. Forstner, 2008. Fluorescence imaging of membrane dynamics. Annu. Rev. Biomed. Eng., 10: 311-338.
- Snapp, E.L., N. Altan and J. Lippincott-Schwartz, 2003. Measuring protein mobility by photobleaching GFP chimeras in living cells. CP Cell Biol., 19.
- Haustein, E. and P. Schwille, 2004. Single-molecule spectroscopic methods. Curr. Opin. Struct. Biol., 14: 531-540.
- Petersen, N.O., P.L. Höddelius, P.W. Wiseman, O. Seger and K.E. Magnusson, 1993. Quantitation of membrane receptor distributions by image correlation spectroscopy: Concept and application. Biophys. J., 65: 1135-1146.
- Hebert, B., S. Costantino and P.W. Wiseman, 2005. Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys. J., 88: 3601-3614.
- Semrau, S. and T. Schmidt, 2007. Particle image correlation spectroscopy (PICS): Retrieving nanometer-scale correlations from high-density single-molecule position data. Biophys. J., 92: 613-621.
- Kraft, M.L., P.K. Weber, M.L. Longo, I.D. Hutcheon and S.G. Boxer, 2006. Phase separation of lipid membranes analyzed with high-resolution secondary ion mass spectrometry. Science, 313: 1948-1951.
- Nägerl, U.V., K.I. Willig, B. Hein, S.W. Hell and T. Bonhoeffer, 2008. Live-cell imaging of dendritic spines by STED microscopy. Proc. Natl. Acad. Sci. U.S.A., 105: 18982-18987.
- Sengupta, P., S.B. van Engelenburg and J. Lippincott-Schwartz, 2014. Superresolution imaging of biological systems using photoactivated localization microscopy. Chem. Rev., 114: 3189-3202.
- Ianoul, A. and L.J. Johnston, 2007. Near-Field Scanning Optical Microscopy to Identify Membrane Microdomains. In: Methods in Membrane Lipids, Dopico, A.M. (Ed.), Humana Press, Totowa, New Jersey, ISBN: 978-1-59745-519-0, pp: 469-480.
- Cai, Y., M.J. Hossain, J.K. Hériché, A.Z. Politi and N. Walther et al., 2018. Experimental and computational framework for a dynamic protein atlas of human cell division. Nature, 561: 411-415.
- Xu, W., Z. Zeng, J.H. Jiang, Y.T. Chang and L. Yuan, 2016. Discerning the chemistry in individual organelles with small-molecule fluorescent probes. Angew. Chem. Int. Edn., 55: 13658-13699.
- Takakura, H., Y. Zhang, R.S. Erdmann, A.D. Thompson and Y. Lin et al., 2017. Long time-lapse nanoscopy with spontaneously blinking membrane probes. Nat. Biotechnol., 35: 773-780.
- Zhang, H., C. Aonbangkhen, E.V. Tarasovetc, E.R. Ballister, D.M. Chenoweth and M.A. Lampson, 2017. Optogenetic control of kinetochore function. Nat. Chem. Biol., 13: 1096-1101.
- Liu, A.P., R.J. Botelho and C.N. Antonescu, 2017. The big and intricate dreams of little organelles: Embracing complexity in the study of membrane traffic. Traffic, 18: 567-579.
- Tiwary, A.K. and Y. Zheng, 2019. Protein phase separation in mitosis. Curr. Opin. Cell Biol., 60: 92-98.
- Rai, A.K., J.X. Chen, M. Selbach and L. Pelkmans, 2018. Kinase-controlled phase transition of membraneless organelles in mitosis. Nature, 559: 211-216.
- Dutta, S., R. Chhabra, V. Muthusamy, N. Gain and R. Subramani et al., 2024. Allelic variation and haplotype diversity of Matrilineal (MTL) gene governing in vivo maternal haploid induction in maize. Physiol. Mol. Biol. Plants, 30: 823-838.
- Dutta, S., R.U. Zunjare, A. Sil, D.C. Mishra and A. Arora et al., 2024. Prediction of matrilineal specific patatin-like protein governing in-vivo maternal haploid induction in maize using support vector machine and di-peptide composition. Amino Acids, 56.
How to Cite this paper?
APA-7 Style
Bhattacharya,
S., Dutta,
S., Chowrasia,
S., ,
S. (2024). Unveiling the Dynamics: A Comprehensive Review of Biological Membrane Kinetics During Cell Division. Asian Journal of Biological Sciences, 17(4), 815-845. https://doi.org/10.3923/ajbs.2024.815.845
ACS Style
Bhattacharya,
S.; Dutta,
S.; Chowrasia,
S.; ,
S. Unveiling the Dynamics: A Comprehensive Review of Biological Membrane Kinetics During Cell Division. Asian J. Biol. Sci 2024, 17, 815-845. https://doi.org/10.3923/ajbs.2024.815.845
AMA Style
Bhattacharya
S, Dutta
S, Chowrasia
S,
S. Unveiling the Dynamics: A Comprehensive Review of Biological Membrane Kinetics During Cell Division. Asian Journal of Biological Sciences. 2024; 17(4): 815-845. https://doi.org/10.3923/ajbs.2024.815.845
Chicago/Turabian Style
Bhattacharya, Sampurna, Suman Dutta, Soni Chowrasia, and Sumanta Das .
2024. "Unveiling the Dynamics: A Comprehensive Review of Biological Membrane Kinetics During Cell Division" Asian Journal of Biological Sciences 17, no. 4: 815-845. https://doi.org/10.3923/ajbs.2024.815.845

This work is licensed under a Creative Commons Attribution 4.0 International License.



