Psychrophilic Amylase and Protease Production Using Aspergillus oryzae CP3
| Received 30 Mar, 2024 |
Accepted 02 May, 2024 |
Published 30 Sep, 2024 |
Background and Objective: Majority of industrial enzymes are produced by fungi. Psychrophilic hydrolytic enzymes are required for cold water detergents, biosensors food additives etc. The aim of present experiments is to produce Psychrophilic Amylase and Protease enzymes by SSF and SMF to get high production yields of both enzymes. Materials And Methods: Aspergillus oryzae CP3 was isolated from food preserved in refrigerators and found to produce amylase and protease. The strain was used for amylase and protease production by SSF and SMF. The SSF was conducted with rice and wheat brawns supplemented with nutrition broth. Results: More amylase is produced (7800 IU/gds) in SSF where as more protease is produced (500 IU/gds) in SMF. Further impact of temperature calcium and chloride ions, specific activity of enzymes was tested and temperature 25̊C, pH 8.5, Ca2+ (100 Mm), Cl‾ (300 mm) were found for the high specific activity of both enzymes. Hence, these enzymes are highly suitable for cold detergents application. Conclusion: The study concludes that the isolated Aspergillus oryzae can be a potent source for amylase and protease production. Amylase and protease produced can be used in cold detergents.
| Copyright © 2024 Sayeed et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Enzymes are produced by living organisms that act as catalyst, accelerate biochemical reactions. Due to their catalytic activity and biological abundance, they play a major role and are used in industrial applications1. Microorganisms are ideal for enzyme production due to their higher multiplication rate2. Majority of the enzymes used for industrial applications include hydrolytic enzymes for the degradation of various natural substances. Proteases are the most used enzymes for industrial applications as they are widely used in detergent and dairy industries. Amylases are the most used enzymes after proteases they have a wide set of applications in baking, starch and textile industries3.
Psychrophilic microorganisms are the group of cold-tolerant microorganisms ranging below 20°C. They usually are cold tolerant and the enzymes produced by these organisms are of greater use due to their high stability and low contamination at lower temperatures making them an optimal choice
for industrial applications. Enzymes produced by psychrophilic organisms are stable at low temperatures whereas the enzymatic activity of enzymes produced by non-psychrophilic organisms is decreased at lower temperatures. The enzymes produced by psychrophilic organisms play a pivotal role in the manufacturing of cold-water detergents, biosensors and food additives4. Enzymes are mainly produced by a process known as fermentation. Fermentation is majorly carried out in two ways for enzyme production which include solid state fermentation (SSF) and submerged fermentation (SMF). The SSF is used for the production of fungal enzymes whereas SMF is used for the production of bacterial enzymes5. The SSF is a beneficial method when working on fungi as it can grow well on solid substrate lesser amount of water required compared to submerged fermentation resulting in cost-efficient production of enzymes hence decreasing the product price6.
The objective of this study was to produce Amylase and Protease from psychrophilic Aspergillus oryzae fungi by submerged and solid-state fermentation and further purifying the enzyme and assessing its properties.
MATERIALS AND METHODS
The study was carried out from June, 2023 to February, 2024 at Department of Microbiology, Nizam College, Basheer Bagh Hyderabad, India
Isolation of fungi producing amylase and protease: Psychrophilic fungal species were isolated from low-temperature preserved bread, milk, fridge coatings and starchy soil. The fungal samples were diluted into 10-4 fold dilutions and 100 μL of samples were spread on PDA, without dextrose. Plates were coated with ringers iodine and strains with more clear zone were selected. The resultant fungal isolates were collected and subcultured. The 5 isolates were selected from screening for amylase-producing fungi. These five fungal isolates were subcultured on skim milk agar plates. The fungi showed good growth and a hydrolytic zone on skim milk agar with amylolytic activity was further used. One fungal isolate was selected for both amylase and protease production.
Colony characteristics: Isolated fungi were observed for colony morphology on PDA agar.
Microscopy: The fungal samples grown on petri plates were identified by lactophenol cotton blue staining. The hyphae are selected with sterile forceps and placed on a clean slide and lactophenol cotton blue is added and observed under a microscope with a cover slip.
Biochemical test
Starch hydrolysis method: The production of amylase in fungi is confirmed by starch hydrolysis method where the amylase-producing fungal plate is flooded with ringers iodine solution containing 3 g of KI and 1 g of iodine in 100 mL of Water and excess solution is removed. The amylase-producing strains of fungi are identified by decolourized zones around the colonies due to amylase production and starch hydrolysis.
Screening for protease activity: Obtained fungal isolates with possible proteolytic activity were screened using skim milk agar. As 10% of non-fat milk is added to 100 mL distilled water and autoclaved at 15psi for 15 min. The isolated fungal samples were inoculated in petri plates and incubated for 3 days at room temperature. The grown fungal colonies form a clear zone around the media indicating the protease production7.
Production of amylase and protease by submerged fermentation: Broth media with peptone (6.0 g/L), MgSO4 (0.5 g/L), KCl (0.5 g/L), starch (6 g/ L), glucose (5 g/L), NH4Cl, peptone and yeast extract (each 2 g/L) is prepared for submerged fermentation. Inoculum was made by inoculating the culture in 30 mL of media in 100 mL flask. The culture was incubated at 28°C on a shaker with 120 rpm for 2 days. As 5% of the inoculum was transferred to production media and incubated for 6 days.
Production of amylase and protease by solid state fermentation: The composition of the fermentation medium (gL–1): Glucose, 20; (NH4)2SO4, 6.6; KH2PO4, 3.5; FeSO4.7H2O, 0.15; MgSO4.7H2O, 0.10; MnCl2.2H2O, 0.45 and peptone, 3.0; at pH 6.8, wheat brawn and rice brawn (1:1) were selected as substrates and air dried and introduced into 250 mL flask containing 30 g of solid media, media broth was added to make up the moisture level to 60%8. The prepared media was autoclaved at 15 psi for 15 min. The 5% inoculum containing mass suspension was added to a sterile fermentation medium and incubated for 6 days at 25°C.
Enzyme assay for amylase and protease
Amylase enzyme assay: The reaction mixture was prepared by combining 1.25 mL of 1% soluble starch, 0.25 mL of 0.1 M acetate buffer (pH 5.0), 0.25 mL of distilled water and 0.25 mL of enzyme. The mixture was then incubated at 30°C for 10 min. Following the incubation period, the reaction was stopped by adding 0.5 mL of DNS reagent. The reaction mixture was subsequently boiled for 5 min to develop color. After boiling, the mixture was allowed to cool and if necessary, it was diluted to ensure that the absorbance fell within the detection range. The absorbance of the solution was measured at 575 nm using a Helico colorimeter, made in Hyderabad, India.
A standard curve was prepared using known concentrations of glucose. The amount of reducing sugars (glucose equivalents) liberated by the enzyme was then calculated using the standard curve. The enzyme activity was determined in units (IU), where one unit was defined as the amount of enzyme that released one μmol of glucose equivalent per min under the assay conditions9.
Protease enzyme assay: The assay was done as 2% of casein solution (1 mL) was incubated with 0.1 mL of enzyme solution and 0.9 mL of sodium phosphate buffer (pH 7) for 10 min at 30°C. The reaction was halted using 10% TCA solution. The mixture was subjected to centrifugation at 10,000 rpm for 5 min. The colour intensity of the supernatant was measured at 280 nm and enzyme activity was calculated from a standard curve of L-tyrosine.
Extraction of enzymes: Dry substrate of SSF was extracted with phosphate buffer (pH 6.8) at 1:50 dilution. Enzyme extraction in crude form was carried out by collecting the culture liquid of SMF and buffer mixed SSF was centrifuged at 10,000 rpm for 10 min. The supernatant was taken leaving the cell containing pellet on the bottom. The cell-free supernatant was filtered through Whatman no 1 filter paper and the filtrate acts as a crude enzyme.
Purification of enzymes
Purification of amylase: Ammonium sulfate (70%) was utilized to fully saturate the supernatant under continuous stirring at 4°C. Subsequently, a 20 min centrifugation was conducted at 14,000×g. The fraction containing ammonium sulfate within a dialysis bag was then dialyzed against 0.05 M sodium phosphate buffer (pH 6.9, 4°C) for 6 hrs. Following this, the concentrated enzyme was applied onto a DEAE-cellulose column, pre-stabilized with the same buffer. Elution of the enzyme was carried out using a similar buffer at a flow rate of 1 mL/min, employing a linear gradient of 0.05-0.5 M NaCl. The resulting fractions were collected and their protein concentration was determined at 280 nm. Fractions exhibiting the highest protein concentration were subsequently assessed for α-amylase activity.
Purification of protease: The culture was collected and made cell-free by centrifuging at 10,000 rpm for 10 min and the supernatant was collected as an enzyme source. The enzyme was precipitated from the
supernatant by the addition of ammonium sulphate of 70% saturation and transferred into a dialysis bag for dialysis. The bag containing enzymes was submerged in 1000 mL of 0.01 M of phosphate buffer and kept for dialysis for 48 hrs, being stirred on a magnetic stirrer. Further purification was done by column chromatography using Diethyl Amino Ethyl (DEAE) anion column10.
Characterization of enzymes: Protein content and protease activity were assessed and specific activity was computed as enzyme activity per millig of protein. Purified enzyme was tested for characterization of enzymes. Various buffers (0.1 M) with pH levels spanning from 4, 5, 6, 7, 7.5, 8, 8.5, 9, 10, 11 and 12 were employed to investigate the impact on enzyme activity. Temperature (15, 20, 25, 30, 35, 40 and 45°C) and ions (Ca2+, Cl–) concentrations were also tested to know optimum conditions for enzyme activity.
SPECIFIC ACTIVITY
For amylase: The specific activity of amylase was investigated. Initially, 10 mg of amylase displayed an activity level of 124 U, serving as the basis for further analysis, 1 mg of amylase exhibited a specific activity of 12.4 U per millig activity.
For protease: The specific activity of Protease, was investigated. Initially, 10 mg of amylase displayed an activity level of 108 U, serving as the basis for further analysis, 1 mg of amylase exhibited a specific activity of 10.8 U per millig activity.
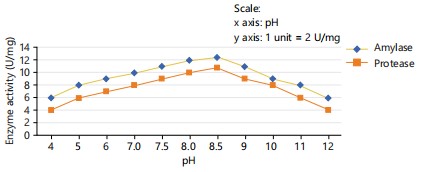
Effect of pH: The impact of initial medium pH on the activity of protease and amylase was investigated. Various initial pH values 4, 5, 6, 7, 7.5, 8, 8.5, 9, 10, 11 and 12 were examined to determine their effect on the activity of these enzymes.
Effect of temperature: The optimal temperature for the activity of protease and amylase enzymes from Aspergillus oryzae is crucial for maximizing enzyme activity. The temperatures (15, 20, 25, 30, 35, 40 and 45°C) had been tested for specific activity of enzymes.
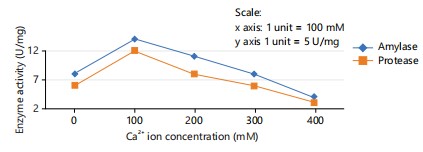
Effect of Ca2+ and Cl–ions: The effect of calcium ions was tested at various concentrations ranging 100, 200, 300 and 400 mM on protease and amylase activity.
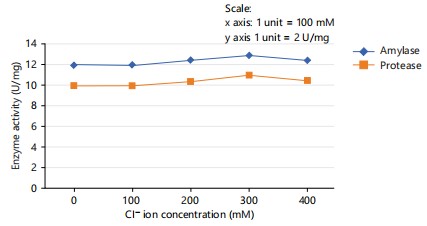
The effect of chloride ions at various concentrations ranging from 100, 200, 300 and 400 mM was investigated to assess the impact of chloride ions on protease and amylase activity, alongside a control group without any ions.
Statistical analysis: Experiments were carried out in triplicates for three times (n = 9). Statistical analysis of the study was done using MS Excel. Values are expressed as Means±SD.
RESULTS
Fungal characteristics and morphology: The colony morphology was observed as yellow green spore-bearing bodies with dense and wooly colonies. Microscopic observations showed large thick-walled conidiophores, the vegetative hyphae of the fungi are septate and the conidia are green and globuse to subglobuse. Isolated fungal sps were identified as Aspergillus oryzae by colony morphology and microscopic observations and named as Aspergillus oryzae CP3.
Biochemical test: The isolated fungi produce both Amylase and Protease enzymes. The presence of Amylase production is identified by the zones formed when the agar plate is flooded with ringers iodine and the decolourized zones indicates the starch hydrolysis and amylolytic activity, the fungal isolates with large zones indicate more amylase production.
The proteolytic activity was also determined for the fungal isolate the fungal samples spread on the skim milk agar showed the zones which indicate the proteolytic activity.
Enzyme production: The enzymes had been produced by A.oryzae. Amylase produced in SSF was 7800 U/gds whereas in SMF 121 μ/mL. The protease in SSF after was 1100 U/gds whereas in SMF 500 μ/mL. In SSF more amylase (7800 U/gds) is produced compared to proteas (1100 U/gds) whereas protease was produced more by SMF (500 IU/mL) compares to amylase(121 IU/mL) the results of enzyme production.
Enzyme purification: The purification steps, comprising the precipitation of ammonium sulfate, dialysis and chromatography were the stages that reported considerable recovery in enzyme activity. The recovery percentages of these enzymes ranged between 61-64% for the two types of enzymes (Table 1 and 2). The data show that critical for the advancement of purification technologies aimed at the increasing output and recovery of these precious enzymes.
Determining the optimal temperature level: Results showed that at 35°C the highest levels of protease and amylase specific activities of 12 and 14 U/mg in SSF, respectively. Additionally, the maximum activity of crude protease and amylase was determined at 35°C. The optimal temperature results are given in Fig. 1.
Even if 35°C is optimal, activity is retained till 15°C, making it suitable to work at low temperatures.
Ca2+ and Cl–: The calcium 100 mM yielded the highest protease and amylase activities, conversely, in the absence of calcium ions, enzyme activity was significantly reduced. These findings underscore the supportive role of calcium ions in maintaining the structural integrity and stability of protease and amylase. The optimal results of Ca2+ ions are given in Fig. 2.
| Table 1: | Solid state fermentation produced enzymes purification | |||
| Method | Amylase | Percentage of recovery (%) |
Protease | Percentage of recovery (%) |
| Crude | 156 IU/gds×150 mL =23400 | 100 | 22 IU/gds×150 mL =3300 | 100 |
| Ammonium sulphate+dialysis | 1765 IU×11 mL =19415 | 82 | 270 IU×10 mL =2700 | 81 |
| DEAE chromatography | 2524 IU×6 mL =15144 | 63.96 | 411 IU×5 mL = 2055 | 61.65 |

|
|
|
|
| Table 2: | Submerged fermentation produced enzymes purification | |||
| Method | Amylase | Percentage of recovery (%) |
Protease | Percentage of recovery (%) |
| Crude | 121 IU×105 mL = 12705 | 100 | 500 IU×105 mL = 5500 | 100 |
| Ammonium sulphate+dialysis | 825 IU×12 mL = 9900 | 77.9 | 3722 IU×11 mL = 40942 | 77.9 |
| DEAE chromatography | 1625 IU×5 mL = 8125 | 63.9 | 5391 IU×6 mL = 32346 | 61.5 |
The results revealed no significant enhancement in protease and amylase activity in the presence and absence of chloride. The presence of 300 mM chloride ion led to a little increase in protease and amylase activity but it is not significant. The optimal result of Cl- ions is given in Fig 3.
Determining the optimal pH level: The results indicated that the enzyme is activity in a broad pH range of 7.0 to 11.0. However, the highest levels of activity yield were achieved when the reaction pH was set at 8.5. Consequently, the optimal pH for the protease and amylase activity was determined to be 8.5. Notably, it was observed that under alkaline conditions, both enzymes exhibited higher activity compared to acidic conditions. The results of pH is given is given in Fig 4.
DISCUSSION
Psychrophilic Aspergillus oryzae (CP3) was isolated producing amylase and protease from cold food samples. The SSF (solid state fermentation) and SMF (submerged fermentation) methods for enzyme production was carried out and compared. Enzymes production was comparatively higher in solid-state fermentation (SSF) than submerged fermentation (SMF). For instance, in amylase production, SSF yielded 7800 U/gds compared to SMF’s 121 U/mL. Similarly, SSF showed better results in protease production, yielding 1100 U/gds compared to SMF’s 500 U/mL. Londoño-Hernández et al.9 found alkaline protease activity to be approximately 23 U/mL in SSF. Belmessikh et al.10 achieved 21309 U/g in SSF and 2343.5 U/g in SMF. Das and Prasad11observed protease activity up to 202.2 U/gds. Muñoz-Seijas et al.12 produced Protease by SSF using inscts frass as raw material. Fadel et al.13reported amylase production of 149.2 U/gds, while Melnichuk et al.14achieved 45,900 U/g of dry substrate in amylase activity. Purification steps, including ammonium sulfate precipitation, dialysis and chromatography, led to considerable recovery in enzyme activity. In SSF, crude amylase and protease extracted were approximately 23,400 and 3300, respectively, after the first stage of ammonium sulfate precipitation, with an 80% recovery after dialysis. In SMF, the crude extractions were around 12705 IU for amylase and 5500 IU for protease, with a 78% recovery after dialysis. From the precipitation with ions, the enzymes showed a recovery percentage of Amylase and Protease is 64 and 62%, respectively in SSF whereas in SMF the protease and amylase enzymes showed a recovery percentage of 64 and 61% indicating higher recovery of amylase compared to protease.
Arunachallam et al.15 observed a yield of 24%. Braia et al.16 achieved a 65% recovery of alpha-amylase. The specific activity of amylase and protease was found to be 12.4 and 10.8 u/mg, respectively. Mamo et al.17observed a specific activity of 0.13758 u/mg for milk-clotting protease. Souza et al.18reported a specific activity of 0.0558 u/mg for protease, while Niyonzima and More19 observed 75 u/mg for protease. Khoo et al.20 noted amylase activity of about 50 U/mg, while Negi and Banerjee21 noted amylase activity around 16 u/mg. The optimal temperature for both protease and amylase was observed at 35°C both significant activity was observed at 15°C, with a pH range of activity between 7.0 and 11.0, particularly showing better reactivity at pH 8.5 alkaline conditions. The presence of Ca2+ activated the enzyme’s activity, while Cl- does not affect activity.
Sadh et al.22observed optimal pH and temperature ranges of 7.4 and 37°C, respectively, for enzymatic reactions utilizing protease produced by Aspergillus oryzae, on the other hand, Amylase and Protease exhibited greater stability at 50°C with a pH of 4.95 and 53.4°C23 and the strain was incubated at 28°C with a pH of 624. The present study implicates the production of Amylase by SSF and protease by SMF. The selected strain can be used for Protease and Amylase production by changing the fermentation conditions. The fermentation conditions provided can be used for other strains also. It is recommended to have a potent single strain for the production of two enzymes at different conditions. Further factors influencing enzyme production should be optimized and enzymes should be produced at a large scale.
CONCLUSION
Psychrophilic amylase and protease were produced by SSF and SMF using isolated Aspergillus oryzae (CP3) strain, from the studies SSF was found to be suitable for amylase production and SMF was found to be suitable for protease production. Ammonium precipitation- dialysis and DEAE anion exchange chromatography were found to be effective in amylase and protease purification. Enzymes activities optimized and found to be suitable for cold detergent applications.
SIGNIFICANCE STATEMENT
For detergents multiple enzymes are required. Psychrophilic enzymes are required in detergents working at room temperature and below. Psychrophilic Aspergillus oryzae CP3 was isolated in the laboratory and used for Amylase and Protease production. More amylase is produced in solid state fermentation whereas more protease is produced in Submerged Fermentation. The study is significant because it produces Amylase and Protease by SSF and SMF by a single fungal strain Aspergillus oryzae CP3.
REFERENCES
- Singh, R., M. Kumar, A. Mittal and P.K. Mehta, 2016. Microbial enzymes: Industrial progress in 21st century. 3 Biotech, 6.
- Dahiya, S., B.K. Bajaj, A. Kumar, S.K. Tiwari and B. Singh, 2020. A review on biotechnological potential of multifarious enzymes in bread making. Process Biochem., 99: 290-306.
- Gurung, N., S. Ray, S. Bose and V. Rai, 2013. A broader view: Microbial enzymes and their relevance in industries, medicine, and beyond. BioMed Res. Int., 2013.
- Banerjee, R., A. Halder and A. Natta, 2016. Psychrophilic microorganisms: Habitats and exploitation potentials. Eur. J. Biotechnol. Biosci., 4: 16-24.
- Ahmed, A., F.U.H. Nasim, K. Batool and A. Bibi, 2017. Microbial β-Glucosidase: Sources, production and applications. J. Appl. Environ. Microbiol., 5: 31-46.
- Bhargav, S., B.P. Panda, M. Ali and S. Javed, 2008. Solid-state fermentation: An Overview. Chem. Biochem. Eng. Q., 22: 49-70.
- Maitig, A.M.A., M.A.M. Alhoot and K. Tiwari, 2018. Isolation and screening of extracellular protease enzyme from fungal isolates of soil. J. Pure Appl. Microbiol., 12: 2059-2068.
- Swargiari, B.N. and P.K. Baruah, 2012. Production of microbial α-amylase by solid state fermentation-An overview. Int. J. Curr. Res., 4: 350-356.
- Londoño-Hernández, L., M. de Jesús García-Gómez, S. Huerta-Ochoa, A.M. Polanía-Rivera, C.N. Aguilar and L.A. Prado-Barragán, 2024. Effect of glucose concentration on the production of proteolytic extract by different strains of Aspergillus under solid-state fermentation. Fermentation, 10.
- Belmessikh, A., H. Boukhalfa, A. Mechakra-Maza, Z. Gheribi-Aoulmi and A. Amrane, 2013. Statistical optimization of culture medium for neutral protease production by Aspergillus oryzae. Comparative study between solid and submerged fermentations on tomato pomace. J. Taiwan Inst. Chem. Eng., 44: 377-385.
- Das, G. and M.P. Prasad, 2010. Isolation, purification & mass production of protease enzyme from Bacillus subtilis. Int. Res. J. Microbiol., 1: 26-31.
- Muñoz-Seijas, N., H. Fernandes, D. Outeiriño, M.G. Morán-Aguilar, J.M. Domínguez and J.M. Salgado, 2024. Potential use of frass from edible insect Tenebrio molitor for proteases production by solid-state fermentation. Food Bioprod. Process., 144: 146-155.
- Fadel, M., S. AbdEl-Halim, H. Sharada, A. Yehia and M. Ammar, 2020. Production of glucoamylase, α-amylase and cellulase by Aspergillus oryzae F-923 cultivated on wheat bran under solid state fermentation. J. Adv. Biol. Biotechnol., 23: 8-22.
- Melnichuk, N., M.J. Braia, P.A. Anselmi, M.R. Meini and D. Romanini, 2020. Valorization of two agroindustrial wastes to produce alpha-amylase enzyme from Aspergillus oryzae by solid-state fermentation. Waste Manage., 106: 155-161.
- Arunachallam, P., V. Kumaravel and S.R. Gopal, 2024. Purification and biochemical characterization of α-amylase from Aspergillus tamarii MTCC5152. Prep. Biochem. Biotechnol., 54: 444-453.
- Braia, M., I. Cabezudo, V.L. Barrera, P. Anselmi, M.R. Meini and D. Romanini, 2021. An optimization approach to the bioconversion of flour mill waste to α-amylase enzyme by Aspergillus oryzae. Process Biochem., 111: 102-108.
- Mamo, J., M. Kangwa, H.M. Fernandez-Lahore and F. Assefa, 2020. Optimization of media composition and growth conditions for production of milk-clotting protease (MCP) from Aspergillus oryzae DRDFS13 under solid-state fermentation. Braz. J. Microbiol., 51: 571-584.
- Souza, P.M., G. Werneck, B. Aliakbarian, F. Siqueira and E.X.F. Filho et al., 2017. Production, purification and characterization of an aspartic protease from Aspergillus foetidus. Food Chem. Toxicol., 109: 1103-1110.
- Niyonzima, F.N. and S.S. More, 2015. Purification and characterization of detergent-compatible protease from Aspergillus terreus gr. 3 Biotech, 5: 61-70.
- Khoo, S.L., A.A. Amirul, M. Kamaruzaman, N. Nazalan and M.N. Azizan, 1994. Purification and characterization of α-amylase from Aspergillus flavus. Folia Microbiol., 39: 392-398.
- Negi, S. and R. Banerjee, 2009. Characterization of amylase and protease produced by Aspergillus awamori in a single bioreactor. Food Res. Int., 42: 443-448.
- Sadh, P.K., S. Duhan and J.S. Duhan, 2018. Agro-industrial wastes and their utilization using solid state fermentation: A review. Bioresour. Bioproces., 5, No. 1.
- Hernandez, M.S., M.R. Rodriguez, N.P. Guerra and R.P. Roses, 2006. Amylase production by Aspergillus niger in submerged cultivation on two wastes from food industries. J. Food Eng., 73: 93-100.
- Salihi, A., A. Asoodeh and M. Aliabadian, 2017. Production and biochemical characterization of an alkaline protease from Aspergillus oryzae CH93. Int. J. Biol. Macromol., 94: 827-835.
How to Cite this paper?
APA-7 Style
Sayeed,
M., Hassaan,
A.B., Mohiuddin,
M., Vanigeri,
M., Bunnawar,
K., Kausar,
Z., Pasha,
C. (2024). Psychrophilic Amylase and Protease Production Using Aspergillus oryzae CP3. Asian Journal of Biological Sciences, 17(3), 388-396. https://doi.org/10.3923/ajbs.2024.388.396
ACS Style
Sayeed,
M.; Hassaan,
A.B.; Mohiuddin,
M.; Vanigeri,
M.; Bunnawar,
K.; Kausar,
Z.; Pasha,
C. Psychrophilic Amylase and Protease Production Using Aspergillus oryzae CP3. Asian J. Biol. Sci 2024, 17, 388-396. https://doi.org/10.3923/ajbs.2024.388.396
AMA Style
Sayeed
M, Hassaan
AB, Mohiuddin
M, Vanigeri
M, Bunnawar
K, Kausar
Z, Pasha
C. Psychrophilic Amylase and Protease Production Using Aspergillus oryzae CP3. Asian Journal of Biological Sciences. 2024; 17(3): 388-396. https://doi.org/10.3923/ajbs.2024.388.396
Chicago/Turabian Style
Sayeed, Mohd, Ahmed Bin Saleh Hassaan, Majid Mohiuddin, Meghana Vanigeri, Keerthi Bunnawar, Zainab Kausar, and Chand Pasha.
2024. "Psychrophilic Amylase and Protease Production Using Aspergillus oryzae CP3" Asian Journal of Biological Sciences 17, no. 3: 388-396. https://doi.org/10.3923/ajbs.2024.388.396

This work is licensed under a Creative Commons Attribution 4.0 International License.


